-Search query
-Search result
Showing all 29 items for (author: gilmore & r)

EMDB-25673: 
Prepore structure of pore-forming toxin Epx1

PDB-7t4e: 
Prepore structure of pore-forming toxin Epx1

EMDB-8926: 
Mutated p53-DNA assembly

EMDB-8927: 
Wild-type p53-DNA assembly

EMDB-4306: 
Subtomogram average of unsorted ribosome-translocon complexes from STT3B(-/-) HEK cells

EMDB-4307: 
Subtomogram average of OST-free ribosome-translocon complexes from STT3B(-/-) HEK cells

EMDB-4308: 
Subtomogram average of OST-containing ribosome-translocon complexes from STT3B(-/-) HEK cells
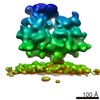
EMDB-4309: 
Subtomogram average of unsorted ribosome-translocon complexes from STT3A(-/-) HEK cells
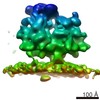
EMDB-4310: 
Subtomogram average of OST-free ribosome-translocon complexes from STT3A(-/-) HEK cells

EMDB-4311: 
Subtomogram average of ribosome-translocon complexes from STT3A(-/-) HEK cells containing an unidentified ER-lumenal component
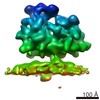
EMDB-4312: 
Subtomogram average of unsorted ribosome-translocon complexes from wild type HEK cells
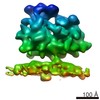
EMDB-4313: 
Subtomogram average of OST-free ribosome-translocon complexes from wild type HEK cells
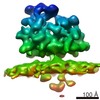
EMDB-4314: 
Subtomogram average of OST-containing ribosome-translocon complexes from wild type HEK cells

EMDB-4315: 
Subtomogram average of OST-containing ribosome-translocon complexes from canine rough microsomal membranes

EMDB-4316: 
Cryo-EM Structure of the Mammalian Oligosaccharyltransferase Bound to Sec61 and the Programmed 80S Ribosome

EMDB-4317: 
Cryo-EM Structure of the Mammalian Oligosaccharyltransferase Bound to Sec61 and the Non-programmed 80S Ribosome

PDB-6ftg: 
Subtomogram average of OST-containing ribosome-translocon complexes from canine rough microsomal membranes

PDB-6fti: 
Cryo-EM Structure of the Mammalian Oligosaccharyltransferase Bound to Sec61 and the Programmed 80S Ribosome

PDB-6ftj: 
Cryo-EM Structure of the Mammalian Oligosaccharyltransferase Bound to Sec61 and the Non-programmed 80S Ribosome
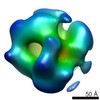
EMDB-6400: 
Electron microscopy of BRCA1(5832insC) mutant-RNAP II transcriptional complex

EMDB-6340: 
Electron cryo-microscopy of a BRCA1-RNAPII transcriptional complex

EMDB-1651: 
Cryo-EM structure of the programmed yeast 80 ribosome bound the Ssh1 complex

EMDB-1667: 
Cryo-EM structure of the active yeast Ssh1 complex bound to the programmed yeast 80S ribosome bearing a P-site tRNA

EMDB-1668: 
Cryo-EM structure of the active yeast 80S ribosome bearing a P-site tRNA and with the rRNA expansion segment ES27 in the exit conformation

EMDB-1669: 
Cryo-EM structures of the idle yeast Ssh1 complex bound to the yeast 80S ribosome

EMDB-1652: 
Cryo-EM structure of the mammalian Sec61 complex bound to the actively translating wheat germ 80S ribosome

PDB-2ww9: 
Cryo-EM structure of the active yeast Ssh1 complex bound to the yeast 80S ribosome

PDB-2wwa: 
Cryo-EM structure of idle yeast Ssh1 complex bound to the yeast 80S ribosome

PDB-2wwb: 
CRYO-EM STRUCTURE OF THE MAMMALIAN SEC61 COMPLEX BOUND TO THE ACTIVELY TRANSLATING WHEAT GERM 80S RIBOSOME
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
