[English] 日本語
 Yorodumi
Yorodumi- PDB-6ftg: Subtomogram average of OST-containing ribosome-translocon complex... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 6ftg | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Subtomogram average of OST-containing ribosome-translocon complexes from canine rough microsomal membranes | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
 Components Components |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
 Keywords Keywords | PROTEIN TRANSPORT / Protein translocon of the endoplasmic reticulum | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationoligosaccharyltransferase complex A / oligosaccharyltransferase complex B / Insertion of tail-anchored proteins into the endoplasmic reticulum membrane / membrane docking / endoplasmic reticulum Sec complex / pronephric nephron development / dolichyl-diphosphooligosaccharide-protein glycotransferase / dolichyl-diphosphooligosaccharide-protein glycotransferase activity / cotranslational protein targeting to membrane / Ssh1 translocon complex ...oligosaccharyltransferase complex A / oligosaccharyltransferase complex B / Insertion of tail-anchored proteins into the endoplasmic reticulum membrane / membrane docking / endoplasmic reticulum Sec complex / pronephric nephron development / dolichyl-diphosphooligosaccharide-protein glycotransferase / dolichyl-diphosphooligosaccharide-protein glycotransferase activity / cotranslational protein targeting to membrane / Ssh1 translocon complex / Sec61 translocon complex / protein insertion into ER membrane / protein-transporting ATPase activity / oligosaccharyltransferase complex / : / protein targeting to ER / SRP-dependent cotranslational protein targeting to membrane, translocation / : / signal sequence binding / : / ribosomal subunit / post-translational protein targeting to membrane, translocation / protein transmembrane transporter activity / ubiquitin ligase inhibitor activity / positive regulation of signal transduction by p53 class mediator / rough endoplasmic reticulum / MDM2/MDM4 family protein binding / post-translational protein modification / cytosolic ribosome / guanyl-nucleotide exchange factor activity / ribosomal large subunit biogenesis / maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / phospholipid binding / regulation of translation / large ribosomal subunit / ribosome biogenesis / ribosome binding / ribosomal large subunit assembly / 5S rRNA binding / large ribosomal subunit rRNA binding / cytosolic large ribosomal subunit / cytoplasmic translation / tRNA binding / rRNA binding / structural constituent of ribosome / ribosome / translation / ribonucleoprotein complex / mRNA binding / synapse / endoplasmic reticulum membrane / nucleolus / endoplasmic reticulum / RNA binding / zinc ion binding / metal ion binding / membrane / nucleus / cytoplasm / cytosol Similarity search - Function | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological species |   | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Method | ELECTRON MICROSCOPY / subtomogram averaging / cryo EM / Resolution: 9.1 Å | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
 Authors Authors | Pfeffer, S. / Foerster, F. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Funding support |  Germany, 3items Germany, 3items
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
 Citation Citation |  Journal: Science / Year: 2018 Journal: Science / Year: 2018Title: Structural basis for coupling protein transport and N-glycosylation at the mammalian endoplasmic reticulum. Authors: Katharina Braunger / Stefan Pfeffer / Shiteshu Shrimal / Reid Gilmore / Otto Berninghausen / Elisabet C Mandon / Thomas Becker / Friedrich Förster / Roland Beckmann /    Abstract: Protein synthesis, transport, and N-glycosylation are coupled at the mammalian endoplasmic reticulum by complex formation of a ribosome, the Sec61 protein-conducting channel, and ...Protein synthesis, transport, and N-glycosylation are coupled at the mammalian endoplasmic reticulum by complex formation of a ribosome, the Sec61 protein-conducting channel, and oligosaccharyltransferase (OST). Here we used different cryo-electron microscopy approaches to determine structures of native and solubilized ribosome-Sec61-OST complexes. A molecular model for the catalytic OST subunit STT3A (staurosporine and temperature sensitive 3A) revealed how it is integrated into the OST and how STT3-paralog specificity for translocon-associated OST is achieved. The OST subunit DC2 was placed at the interface between Sec61 and STT3A, where it acts as a versatile module for recruitment of STT3A-containing OST to the ribosome-Sec61 complex. This detailed structural view on the molecular architecture of the cotranslational machinery for N-glycosylation provides the basis for a mechanistic understanding of glycoprotein biogenesis at the endoplasmic reticulum. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  6ftg.cif.gz 6ftg.cif.gz | 3.7 MB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb6ftg.ent.gz pdb6ftg.ent.gz | Display |  PDB format PDB format | |
| PDBx/mmJSON format |  6ftg.json.gz 6ftg.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/ft/6ftg https://data.pdbj.org/pub/pdb/validation_reports/ft/6ftg ftp://data.pdbj.org/pub/pdb/validation_reports/ft/6ftg ftp://data.pdbj.org/pub/pdb/validation_reports/ft/6ftg | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  4315MC  4306C  4307C  4308C  4309C  4310C  4311C  4312C  4313C 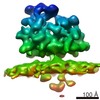 4314C  4316C  4317C  6ftiC  6ftjC C: citing same article ( M: map data used to model this data |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 |
|
- Components
Components
+Protein , 31 types, 31 molecules ABFGHLOPQRSTUVXacdefghkmors2368
-Ribosomal protein ... , 10 types, 10 molecules CIJMNWYjpt
| #3: Protein | Mass: 41115.508 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
|---|---|
| #9: Protein | Mass: 24511.861 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
| #10: Protein | Mass: 19270.318 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
| #12: Protein | Mass: 16217.355 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
| #13: Protein | Mass: 24076.088 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
| #22: Protein | Mass: 7512.774 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
| #24: Protein | Mass: 15891.787 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
| #35: Protein | Mass: 10090.891 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
| #41: Protein | Mass: 10168.153 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
| #44: Protein | Mass: 17689.461 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
-60S ribosomal protein ... , 6 types, 6 molecules DEZbin
| #4: Protein | Mass: 34006.312 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
|---|---|
| #5: Protein | Mass: 28529.848 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
| #25: Protein | Mass: 15704.635 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
| #27: Protein | Mass: 8706.283 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
| #34: Protein | Mass: 11888.371 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
| #39: Protein/peptide | Mass: 3213.075 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
-Protein/peptide , 3 types, 3 molecules l47
| #37: Protein/peptide | Mass: 6295.562 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
|---|---|
| #54: Protein/peptide | Mass: 3809.582 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
| #57: Protein/peptide | Mass: 2145.636 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
-RNA chain , 3 types, 3 molecules uvw
| #45: RNA chain | Mass: 1186579.500 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
|---|---|
| #46: RNA chain | Mass: 38691.914 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
| #47: RNA chain | Mass: 50143.648 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
-Protein transport protein Sec61 subunit ... , 3 types, 3 molecules xyz
| #48: Protein | Mass: 50758.504 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
|---|---|
| #49: Protein | Mass: 7019.456 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
| #50: Protein/peptide | Mass: 3265.008 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
-Dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit ... , 2 types, 2 molecules 15
| #51: Protein | Mass: 14515.871 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
|---|---|
| #55: Protein | Mass: 80657.375 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  References: UniProt: F1PJP5, dolichyl-diphosphooligosaccharide-protein glycotransferase |
-Sugars , 1 types, 1 molecules
| #59: Polysaccharide | alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D- ...alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-3)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose Source method: isolated from a genetically manipulated source |
|---|
-Non-polymers , 3 types, 165 molecules 




| #60: Chemical | ChemComp-MG / #61: Chemical | ChemComp-ZN / #62: Chemical | ChemComp-9UB / [( | |
|---|
-Details
| Has protein modification | Y |
|---|
-Experimental details
-Experiment
| Experiment | Method: ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: PARTICLE / 3D reconstruction method: subtomogram averaging |
- Sample preparation
Sample preparation
| Component |
| ||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Source (natural) |
| ||||||||||||||||||||||||||||||
| Buffer solution | pH: 7.6 | ||||||||||||||||||||||||||||||
| Specimen | Conc.: 2 mg/ml / Embedding applied: NO / Shadowing applied: NO / Staining applied: NO / Vitrification applied: YES | ||||||||||||||||||||||||||||||
| Specimen support | Grid material: MOLYBDENUM | ||||||||||||||||||||||||||||||
| Vitrification | Instrument: FEI VITROBOT MARK IV / Cryogen name: ETHANE-PROPANE / Humidity: 70 % / Details: Blot 3 seconds before plunging. |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
|---|---|
| Microscopy | Model: FEI TITAN KRIOS |
| Electron gun | Electron source:  FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM |
| Electron lens | Mode: BRIGHT FIELD / Nominal defocus max: 4000 nm / Nominal defocus min: 3000 nm |
| Specimen holder | Cryogen: NITROGEN / Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Image recording | Electron dose: 1.5 e/Å2 / Detector mode: COUNTING / Film or detector model: GATAN K2 SUMMIT (4k x 4k) |
- Processing
Processing
| EM software |
| |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CTF correction | Details: On each individual tilt image / Type: PHASE FLIPPING ONLY | |||||||||||||||||||||
| Symmetry | Point symmetry: C1 (asymmetric) | |||||||||||||||||||||
| 3D reconstruction | Resolution: 9.1 Å / Resolution method: FSC 0.5 CUT-OFF / Num. of particles: 17600 / Algorithm: BACK PROJECTION / Symmetry type: POINT | |||||||||||||||||||||
| EM volume selection | Method: Template matching / Num. of tomograms: 211 / Num. of volumes extracted: 27500 | |||||||||||||||||||||
| Atomic model building | B value: 500 / Protocol: RIGID BODY FIT / Space: REAL / Target criteria: Cross-correlation coefficient |
 Movie
Movie Controller
Controller












 PDBj
PDBj
































