[English] 日本語
 Yorodumi
Yorodumi- EMDB-6400: Electron microscopy of BRCA1(5832insC) mutant-RNAP II transcripti... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-6400 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Electron microscopy of BRCA1(5832insC) mutant-RNAP II transcriptional complex | |||||||||
 Map data Map data | Reconstruction of BRCA1-RNAP II mutant complex | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | transcription / DNA damage repair / breast cancer / protein | |||||||||
| Function / homology |  Function and homology information Function and homology informationnegative regulation of mRNA 3'-end processing / histone H2AK127 ubiquitin ligase activity / histone H2AK129 ubiquitin ligase activity / Defective DNA double strand break response due to BRCA1 loss of function / Defective DNA double strand break response due to BARD1 loss of function / BRCA1-BARD1 complex / BRCA1-B complex / BRCA1-C complex / BRCA1-A complex / microfibril binding ...negative regulation of mRNA 3'-end processing / histone H2AK127 ubiquitin ligase activity / histone H2AK129 ubiquitin ligase activity / Defective DNA double strand break response due to BRCA1 loss of function / Defective DNA double strand break response due to BARD1 loss of function / BRCA1-BARD1 complex / BRCA1-B complex / BRCA1-C complex / BRCA1-A complex / microfibril binding / sex-chromosome dosage compensation / negative regulation of centriole replication / random inactivation of X chromosome / ubiquitin-modified histone reader activity / chordate embryonic development / gamma-tubulin ring complex / cellular response to indole-3-methanol / negative regulation of intracellular estrogen receptor signaling pathway / regulation of phosphorylation / nuclear ubiquitin ligase complex / DNA strand resection involved in replication fork processing / homologous recombination / negative regulation of fatty acid biosynthetic process / tissue homeostasis / Regulation of MITF-M-dependent genes involved in DNA replication, damage repair and senescence / protein K6-linked ubiquitination / lateral element / regulation of DNA damage checkpoint / mitotic G2/M transition checkpoint / Impaired BRCA2 binding to PALB2 / negative regulation of protein export from nucleus / XY body / RNA polymerase binding / DNA repair complex / DNA damage tolerance / centrosome cycle / Abortive elongation of HIV-1 transcript in the absence of Tat / FGFR2 alternative splicing / MicroRNA (miRNA) biogenesis / Viral Messenger RNA Synthesis / intracellular membraneless organelle / Signaling by FGFR2 IIIa TM / Homologous DNA Pairing and Strand Exchange / Defective homologous recombination repair (HRR) due to BRCA1 loss of function / Defective HDR through Homologous Recombination Repair (HRR) due to PALB2 loss of BRCA1 binding function / Defective HDR through Homologous Recombination Repair (HRR) due to PALB2 loss of BRCA2/RAD51/RAD51C binding function / Resolution of D-loop Structures through Synthesis-Dependent Strand Annealing (SDSA) / Resolution of D-loop Structures through Holliday Junction Intermediates / HDR through Single Strand Annealing (SSA) / response to ionizing radiation / RNA Pol II CTD phosphorylation and interaction with CE during HIV infection / RNA Pol II CTD phosphorylation and interaction with CE / negative regulation of gene expression via chromosomal CpG island methylation / Impaired BRCA2 binding to RAD51 / Formation of the Early Elongation Complex / Formation of the HIV-1 Early Elongation Complex / mRNA Capping / HIV Transcription Initiation / RNA Polymerase II HIV Promoter Escape / Transcription of the HIV genome / RNA Polymerase II Promoter Escape / RNA Polymerase II Transcription Pre-Initiation And Promoter Opening / RNA Polymerase II Transcription Initiation / RNA Polymerase II Transcription Initiation And Promoter Clearance / mitotic G2 DNA damage checkpoint signaling / Transcriptional Regulation by E2F6 / mRNA Splicing - Minor Pathway / PIWI-interacting RNA (piRNA) biogenesis / negative regulation of cell cycle / negative regulation of reactive oxygen species metabolic process / Processing of Capped Intron-Containing Pre-mRNA / Pausing and recovery of Tat-mediated HIV elongation / Tat-mediated HIV elongation arrest and recovery / Presynaptic phase of homologous DNA pairing and strand exchange / RNA polymerase II transcribes snRNA genes / positive regulation of vascular endothelial growth factor production / HIV elongation arrest and recovery / Pausing and recovery of HIV elongation / ubiquitin ligase complex / Tat-mediated elongation of the HIV-1 transcript / Formation of HIV-1 elongation complex containing HIV-1 Tat / regulation of DNA repair / SUMOylation of DNA damage response and repair proteins / negative regulation of extrinsic apoptotic signaling pathway via death domain receptors / Formation of HIV elongation complex in the absence of HIV Tat / RNA polymerase II, core complex / protein autoubiquitination / RNA Polymerase II Transcription Elongation / Formation of RNA Pol II elongation complex / Maturation of protein E / Maturation of protein E / RNA Polymerase II Pre-transcription Events / ER Quality Control Compartment (ERQC) / Myoclonic epilepsy of Lafora / FLT3 signaling by CBL mutants / mRNA Splicing - Major Pathway / Constitutive Signaling by NOTCH1 HD Domain Mutants / IRAK2 mediated activation of TAK1 complex / Prevention of phagosomal-lysosomal fusion / Alpha-protein kinase 1 signaling pathway Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / negative staining / Resolution: 26.0 Å | |||||||||
 Authors Authors | Winton CE / Gilmore BL / Demmert AC / Tanner JR / Bowman S / Karageorge V / Patel K / Sheng Z / Kelly DF | |||||||||
 Citation Citation |  Journal: Nat Struct Biol / Year: 2001 Journal: Nat Struct Biol / Year: 2001Title: Crystal structure of the BRCT repeat region from the breast cancer-associated protein BRCA1. Authors: R S Williams / R Green / J N Glover /  Abstract: The C-terminal BRCT region of BRCA1 is essential for its DNA repair, transcriptional regulation and tumor suppressor functions. Here we determine the crystal structure of the BRCT domain of human ...The C-terminal BRCT region of BRCA1 is essential for its DNA repair, transcriptional regulation and tumor suppressor functions. Here we determine the crystal structure of the BRCT domain of human BRCA1 at 2.5 A resolution. The domain contains two BRCT repeats that adopt similar structures and are packed together in a head-to-tail arrangement. Cancer-causing missense mutations occur at the interface between the two repeats and destabilize the structure. The manner by which the two BRCT repeats interact in BRCA1 may represent a general mode of interaction between homologous domains within proteins that interact to regulate the cellular response to DNA damage. The structure provides a basis to predict the structural consequences of uncharacterized BRCA1 mutations. #3:  Journal: NAT.STRUCT.BIOL. / Year: 2001 Journal: NAT.STRUCT.BIOL. / Year: 2001Title: Structure of a BRCA1-BARD1 heterodimeric RING-RING complex Authors: Brzovic PS / Rajagopal P / Hoyt DW / King MC / Klevit RE #4:  Journal: J.MOL.BIOL. / Year: 1987 Journal: J.MOL.BIOL. / Year: 1987Title: Structure of ubiquitin refined at 1.8 A resolution Authors: Vijay-Kumar S / Bugg CE / Cook WJ | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_6400.map.gz emd_6400.map.gz | 6.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-6400-v30.xml emd-6400-v30.xml emd-6400.xml emd-6400.xml | 21.7 KB 21.7 KB | Display Display |  EMDB header EMDB header |
| Images |  400_6400.gif 400_6400.gif 80_6400.gif 80_6400.gif | 61.5 KB 14.3 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-6400 http://ftp.pdbj.org/pub/emdb/structures/EMD-6400 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6400 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6400 | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_6400.map.gz / Format: CCP4 / Size: 12.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_6400.map.gz / Format: CCP4 / Size: 12.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Reconstruction of BRCA1-RNAP II mutant complex | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 4 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
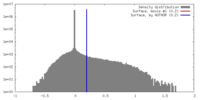
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Mutant BRCA1(5382insC)-RNAP II transcriptional assemblies isolate...
| Entire | Name: Mutant BRCA1(5382insC)-RNAP II transcriptional assemblies isolated from hereditary breast cancer cells |
|---|---|
| Components |
|
-Supramolecule #1000: Mutant BRCA1(5382insC)-RNAP II transcriptional assemblies isolate...
| Supramolecule | Name: Mutant BRCA1(5382insC)-RNAP II transcriptional assemblies isolated from hereditary breast cancer cells type: sample / ID: 1000 Oligomeric state: one mutant BRCA1(5382insC) molecule binds to one polymerase complex Number unique components: 4 |
|---|---|
| Molecular weight | Theoretical: 800 KDa |
-Macromolecule #1: RNA Polyermase II core
| Macromolecule | Name: RNA Polyermase II core / type: protein_or_peptide / ID: 1 / Name.synonym: Pol2 / Number of copies: 1 / Recombinant expression: No / Database: NCBI |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / synonym: human / Tissue: breast / Cell: tumor / Organelle: nucleus Homo sapiens (human) / synonym: human / Tissue: breast / Cell: tumor / Organelle: nucleus |
| Molecular weight | Theoretical: 500 KDa |
| Sequence | UniProtKB: DNA-directed RNA polymerase II subunit RPB1 |
-Macromolecule #2: BRCA1
| Macromolecule | Name: BRCA1 / type: protein_or_peptide / ID: 2 Details: Mutated BRCA1 was attached to tunable microchips using antibodies against the BRCA1 C-terminal domain. Number of copies: 1 / Oligomeric state: monomer / Recombinant expression: No / Database: NCBI |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / synonym: human / Tissue: breast / Cell: tumor / Organelle: nucleus Homo sapiens (human) / synonym: human / Tissue: breast / Cell: tumor / Organelle: nucleus |
| Molecular weight | Theoretical: 200 KDa |
| Sequence | UniProtKB: Breast cancer type 1 susceptibility protein |
-Macromolecule #3: BARD1
| Macromolecule | Name: BARD1 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Oligomeric state: monomer / Recombinant expression: No / Database: NCBI |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / synonym: human / Tissue: breast / Cell: tumor / Organelle: nucleus Homo sapiens (human) / synonym: human / Tissue: breast / Cell: tumor / Organelle: nucleus |
| Molecular weight | Theoretical: 87 KDa |
| Sequence | UniProtKB: BRCA1-associated RING domain protein 1 |
-Macromolecule #4: Ubiquitin
| Macromolecule | Name: Ubiquitin / type: protein_or_peptide / ID: 4 / Number of copies: 2 / Oligomeric state: monomer / Recombinant expression: No / Database: NCBI |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / synonym: human / Tissue: breast / Cell: tumor / Organelle: nucleus Homo sapiens (human) / synonym: human / Tissue: breast / Cell: tumor / Organelle: nucleus |
| Molecular weight | Theoretical: 8 KDa |
| Sequence | UniProtKB: Polyubiquitin-C |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 / Details: 50 mM HEPES, 150 mM NaCl, 10 mM MgCl2, 10 mM CaCl2 |
|---|---|
| Staining | Type: NEGATIVE Details: Protein complexes were adsorbed to antibody-decorated microchips for 2 minutes and stained with 1% uranyl formate for 1 minute. |
| Grid | Details: SiN microchips with TEM windows coated with 25% Ni-NTA functionalized lipid monolayers |
| Vitrification | Cryogen name: NONE / Instrument: OTHER |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI SPIRIT |
|---|---|
| Temperature | Min: 83 K |
| Alignment procedure | Legacy - Astigmatism: Objective lens astigmatism was corrected at high magnification. |
| Details | low-dose illumination |
| Date | Mar 23, 2015 |
| Image recording | Category: CCD / Film or detector model: FEI EAGLE (2k x 2k) / Digitization - Sampling interval: 30 µm / Number real images: 129 / Average electron dose: 5 e/Å2 |
| Electron beam | Acceleration voltage: 120 kV / Electron source: LAB6 |
| Electron optics | Calibrated magnification: 75000 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2 mm / Nominal defocus max: -3.0 µm / Nominal defocus min: -1.0 µm / Nominal magnification: 68000 |
| Sample stage | Specimen holder model: GATAN LIQUID NITROGEN |
| Experimental equipment |  Model: Tecnai Spirit / Image courtesy: FEI Company |
- Image processing
Image processing
| Details | The particles were selected using an automatic selection program. |
|---|---|
| Final reconstruction | Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 26.0 Å / Resolution method: OTHER / Software - Name: RELION / Number images used: 3000 |
| Final two d classification | Number classes: 1 |
-Atomic model buiding 1
| Initial model | PDB ID: Chain - #0 - Chain ID: A / Chain - #1 - Chain ID: B / Chain - #2 - Chain ID: C / Chain - #3 - Chain ID: D / Chain - #4 - Chain ID: E / Chain - #5 - Chain ID: F / Chain - #6 - Chain ID: G / Chain - #7 - Chain ID: H / Chain - #8 - Chain ID: I / Chain - #9 - Chain ID: J / Chain - #10 - Chain ID: K / Chain - #11 - Chain ID: L |
|---|---|
| Software | Name:  Chimera Chimera |
| Details | The domains were separately fitted by manual docking using the Chimera software package. |
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
-Atomic model buiding 2
| Initial model | PDB ID: Chain - Chain ID: A |
|---|---|
| Software | Name:  Chimera Chimera |
| Details | The domains were separately fitted by manual docking using the Chimera software package. |
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
-Atomic model buiding 3
| Initial model | PDB ID: Chain - #0 - Chain ID: A / Chain - #1 - Chain ID: B |
|---|---|
| Software | Name:  Chimera Chimera |
| Details | The domains were separately fitted by manual docking using the Chimera software package. |
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
-Atomic model buiding 4
| Initial model | PDB ID: Chain - Chain ID: A |
|---|---|
| Software | Name:  Chimera Chimera |
| Details | This domain was used to generate a homology based model using the program SWISS-MODEL. |
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
 Movie
Movie Controller
Controller









































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)