-Search query
-Search result
Showing 1 - 50 of 58 items for (author: eisenstein & f)
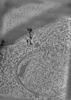
EMDB-38295: 
SPACEtomo example tomograms of Baker's Yeast
Method: electron tomography / : Eisenstein F, Danev R

EMDB-42371: 
Mouse apoferritin imaged with a square aperture
Method: single particle / : Chua EYD, Alink LM, Kopylov M, Johnston J, Einsenstein F, de Marco A

EMDB-42372: 
Mouse apoferritin imaged with a round aperture
Method: single particle / : Chua EYD, Alink LM, Kopylov M, Johnston J, Einsenstein F, de Marco A

EMDB-42373: 
Mouse apoferritin imaged with a square aperture with P2 projection lens rotation, reconstructed from 25,000 particles
Method: single particle / : Chua EYD, Alink LM, Kopylov M, Johnston J, Einsenstein F, de Marco A

EMDB-42374: 
Mouse apoferritin imaged with a round aperture without P2 projection lens rotation, reconstructed from 25,000 particles
Method: single particle / : Chua EYD, Alink LM, Kopylov M, Johnston J, Einsenstein F, de Marco A

EMDB-42851: 
5x5 tiled montage tomogram of a holey carbon grid with apoferritin imaged with a square electron beam
Method: electron tomography / : Chua EYD, Alink LM, Kopylov M, Johnston J, Einsenstein F, de Marco A

EMDB-42879: 
3x3 tiled montage tomogram of a yeast lamella imaged with a square electron beam
Method: electron tomography / : Chua EYD, Alink LM, Kopylov M, Johnston J, Einsenstein F, de Marco A

EMDB-33833: 
In situ subtomogram average of f-actin
Method: subtomogram averaging / : Eisenstein F, Danev R

EMDB-33834: 
Subtomogram average of 70S ribosome (11x11)
Method: subtomogram averaging / : Eisenstein F, Danev R

EMDB-33115: 
Subtomogram average of 70S ribosome
Method: subtomogram averaging / : Eisenstein F, Danev R

EMDB-33116: 
Subtomogram average of 70S ribosome (50S aligned)
Method: subtomogram averaging / : Eisenstein F, Danev R

EMDB-33117: 
Subtomogram average of 70S ribosome (30S aligned)
Method: subtomogram averaging / : Eisenstein F, Danev R

EMDB-33118: 
In situ subtomogram average of 80S ribosome
Method: subtomogram averaging / : Eisenstein F, Danev R

EMDB-12029: 
Cryo-EM structure of the contractile injection system base plate from Anabaena PCC7120
Method: single particle / : Eisenstein F, Weiss GL, Pilhofer M

EMDB-12030: 
Cryo-EM structure of the contractile injection system base plate from Anabaena PCC7120 (C6)
Method: single particle / : Eisenstein F, Weiss GL, Pilhofer M

EMDB-12031: 
Cryo-EM structure of the contractile injection system base plate connector from Anabaena PCC7120
Method: single particle / : Eisenstein F, Weiss GL, Pilhofer M

EMDB-12032: 
Cryo-EM structure of the contractile injection system crown trimer from Anabaena PCC7120
Method: single particle / : Eisenstein F, Weiss GL, Pilhofer M

EMDB-12033: 
Cryo-EM structure of the contractile injection system base plate in contracted form from Anabaena PCC7120
Method: single particle / : Eisenstein F, Weiss GL, Pilhofer M

EMDB-12034: 
Cryo-EM structure of the contractile injection system cap complex from Anabaena PCC7120
Method: single particle / : Eisenstein F, Weiss GL, Pilhofer M

EMDB-13770: 
Cryo-electron tomogram of cryo-FIB milled Anabaena PCC 7120
Method: electron tomography / : Weiss GL, Eisenstein F, Pilhofer M

EMDB-13771: 
Cryo-electron tomogram of cryo-FIB milled Anabaena PCC 7120, cell periphery
Method: electron tomography / : Weiss GL, Eisenstein F, Pilhofer M

EMDB-13772: 
Cryo-electron tomogram of cryo-FIB milled Anabaena PCC 7120 ghost cell
Method: electron tomography / : Weiss GL, Eisenstein F, Pilhofer M

EMDB-13773: 
Subtomogram average of baseplate region of contractile injection systems in Anabaena PCC 7120 in extended state
Method: subtomogram averaging / : Weiss GL, Eisenstein F, Pilhofer M

EMDB-13774: 
Subtomogram average of baseplate region of contractile injection systems in Anabaena PCC 7120 ghost cells in extended state
Method: subtomogram averaging / : Weiss GL, Eisenstein F, Pilhofer M

EMDB-13775: 
Subtomogram average of baseplate region of contractile injection systems in Anabaena PCC 7120 ghost cells in contracted state
Method: subtomogram averaging / : Weiss GL, Eisenstein F, Pilhofer M

PDB-7b5h: 
Cryo-EM structure of the contractile injection system base plate from Anabaena PCC7120
Method: single particle / : Eisenstein F, Weiss GL, Pilhofer M

PDB-7b5i: 
Cryo-EM structure of the contractile injection system cap complex from Anabaena PCC7120
Method: single particle / : Eisenstein F, Weiss GL, Pilhofer M

EMDB-11745: 
Cryo-EM structure of an extracellular contractile injection system in marine bacterium Algoriphagus machipongonensis, the baseplate complex in extended state applied 3-fold symmetry.
Method: single particle / : Xu J, Ericson C

PDB-7aef: 
Cryo-EM structure of an extracellular contractile injection system in marine bacterium Algoriphagus machipongonensis, the baseplate complex in extended state applied 3-fold symmetry.
Method: single particle / : Xu J, Ericson C, Feldmueller M, Lien YW, Pilhofer M

EMDB-11734: 
Cryo-EM structure of an extracellular contractile injection system in marine bacterium Algoriphagus machipongonensis, the cap portion in extended state.
Method: single particle / : Xu J, Ericson C

EMDB-11735: 
Cryo-EM structure of an extracellular contractile injection system in marine bacterium Algoriphagus machipongonensis, the sheath-tube module in extended state.
Method: helical / : Xu J, Ericson C

EMDB-11743: 
Cryo-EM structure of an extracellular contractile injection system in marine bacterium Algoriphagus machipongonensis, the baseplate complex in extended state applied 6-fold symmetry.
Method: single particle / : Xu J, Ericson C

EMDB-11744: 
Focused refined cryo-EM structure of an extracellular contractile injection system in marine bacterium Algoriphagus machipongonensis, the baseplate complex in extended state applied 6-fold symmetry
Method: single particle / : Xu J, Ericson C, Feldmueller M, Lien YW, Pilhofer M

EMDB-11746: 
Cryo-EM structure of the overall extracellular contractile injection system in marine bacterium Algoriphagus machipongonensis
Method: single particle / : Xu J, Ericson C, Feldmueller M, Lien YW, Pilhofer M

EMDB-11747: 
Cryo-EM structure of an extracellular contractile injection system in marine bacterium Algoriphagus machipongonensis, the contracted sheath shell.
Method: helical / : Xu J, Ericson C

EMDB-11748: 
Cryo-EM structure of an extracellular contractile injection system in marine bacterium Algoriphagus machipongonensis, the baseplate complex in contracted state.
Method: single particle / : Xu J, Ericson C, Feldmueller M, Lien YW, Pilhofer M

EMDB-11749: 
sub-tomogram averaging of an extracellular contractile injection system in marine bacterium Algoriphagus machipongonensis
Method: subtomogram averaging / : Xu J, Ericson C, Feldmueller M, Lien YW, Pilhofer M

EMDB-11750: 
sub-tomogram averaging of an extracellular contractile injection system mutant (effector deficient mutant) in marine bacterium Algoriphagus machipongonensis
Method: subtomogram averaging / : Xu J, Ericson C, Feldmueller M, Lien YW, Pilhofer M

EMDB-13704: 
cryo electron tomogram of purified AlgoCIS delta Alg16B
Method: electron tomography / : Xu J, Ericson C, Feldmueller M, Lien YW, Eisenstein F, Pilhofer M

EMDB-13705: 
cryo electron tomogram of Algoriphagus machipongonensis in bacterial late stage
Method: electron tomography / : Xu J, Ericson C, Feldmueller M, Lien YW, Eisenstein F, Pilhofer M

EMDB-13719: 
Cryo electron tomogram of purified AlgoCIS Cgo1/2 deficient mutant
Method: electron tomography / : Xu J, Ericson C, Feldmueller M, Lien YW, Eisenstein F, Pilhofer M

EMDB-13720: 
Cryo electron tomogram of purified AlgoCIS Cgo1 deficient mutant
Method: electron tomography / : Xu J, Ericson C, Feldmueller M, Lien YW, Eisenstein F, Pilhofer M

EMDB-13721: 
Cryo electron tomogram of purified AlgoCIS Cgo2 deficient mutant
Method: electron tomography / : Xu J, Ericson C, Feldmueller M, Lien YW, Eisenstein F, Pilhofer M

EMDB-13722: 
Cryo electron tomogram of purified AlgoCIS wild-type.
Method: electron tomography / : Xu J, Ericson C, Feldmueller M, Lien YW, Eisenstein F, Pilhofer M

EMDB-13723: 
Cryo electron sub-tomogram averaging of purified AlgoCIS delta-Alg16B mutant
Method: subtomogram averaging / : Xu J, Ericson C, Feldmueller M, Lien YW, Eisenstein F, Pilhofer M

EMDB-13724: 
Cryo electron sub-tomogram average of MAC arrays baseplate
Method: subtomogram averaging / : Xu J, Ericson C, Feldmueller M, Lien YW, Eisenstein F, Pilhofer M

EMDB-13725: 
Cryo electron sub-tomogram average of baseplate in Amoebophilus asiaticus
Method: subtomogram averaging / : Xu J, Ericson C, Feldmueller M, Lien YW, Eisenstein F, Pilhofer M

PDB-7adz: 
Cryo-EM structure of an extracellular contractile injection system in marine bacterium Algoriphagus machipongonensis, the cap portion in extended state.
Method: single particle / : Xu J, Ericson C, Feldmueller M, Lien YW, Pilhofer M

PDB-7ae0: 
Cryo-EM structure of an extracellular contractile injection system in marine bacterium Algoriphagus machipongonensis, the sheath-tube module in extended state.
Method: helical / : Xu J, Ericson C, Feldmueller M, Lien YW, Pilhofer M

PDB-7aeb: 
Cryo-EM structure of an extracellular contractile injection system in marine bacterium Algoriphagus machipongonensis, the baseplate complex in extended state applied 6-fold symmetry.
Method: single particle / : Xu J, Ericson C, Feldmueller M, Lien YW, Pilhofer M
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
