[English] 日本語
 Yorodumi
Yorodumi- EMDB-11734: Cryo-EM structure of an extracellular contractile injection syste... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-11734 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of an extracellular contractile injection system in marine bacterium Algoriphagus machipongonensis, the cap portion in extended state. | ||||||||||||
 Map data Map data | |||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | extracellular contractile injection system / STRUCTURAL PROTEIN | ||||||||||||
| Function / homology |  Function and homology information Function and homology information | ||||||||||||
| Biological species |  Algoriphagus machipongonensis (bacteria) Algoriphagus machipongonensis (bacteria) | ||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.5 Å | ||||||||||||
 Authors Authors | Xu J / Ericson C | ||||||||||||
| Funding support |  Switzerland, 3 items Switzerland, 3 items
| ||||||||||||
 Citation Citation |  Journal: Nat Microbiol / Year: 2022 Journal: Nat Microbiol / Year: 2022Title: Identification and structure of an extracellular contractile injection system from the marine bacterium Algoriphagus machipongonensis. Authors: Jingwei Xu / Charles F Ericson / Yun-Wei Lien / Florentine U N Rutaganira / Fabian Eisenstein / Miki Feldmüller / Nicole King / Martin Pilhofer /    Abstract: Contractile injection systems (CISs) are phage tail-like nanomachines, mediating bacterial cell-cell interactions as either type VI secretion systems (T6SSs) or extracellular CISs (eCISs). ...Contractile injection systems (CISs) are phage tail-like nanomachines, mediating bacterial cell-cell interactions as either type VI secretion systems (T6SSs) or extracellular CISs (eCISs). Bioinformatic studies uncovered a phylogenetic group of hundreds of putative CIS gene clusters that are highly diverse and widespread; however, only four systems have been characterized. Here we studied a putative CIS gene cluster in the marine bacterium Algoriphagus machipongonensis. Using an integrative approach, we show that the system is compatible with an eCIS mode of action. Our cryo-electron microscopy structure revealed several features that differ from those seen in other CISs: a 'cap adaptor' located at the distal end, a 'plug' exposed to the tube lumen, and a 'cage' formed by massive extensions of the baseplate. These elements are conserved in other CISs, and our genetic tools identified that they are required for assembly, cargo loading and function. Furthermore, our atomic model highlights specific evolutionary hotspots and will serve as a framework for understanding and re-engineering CISs. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_11734.map.gz emd_11734.map.gz | 30.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-11734-v30.xml emd-11734-v30.xml emd-11734.xml emd-11734.xml | 13.9 KB 13.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_11734.png emd_11734.png | 71 KB | ||
| Filedesc metadata |  emd-11734.cif.gz emd-11734.cif.gz | 5.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-11734 http://ftp.pdbj.org/pub/emdb/structures/EMD-11734 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11734 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11734 | HTTPS FTP |
-Related structure data
| Related structure data |  7adzMC  7ae0C  7aebC  7aefC  7aekC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_11734.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_11734.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.1 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
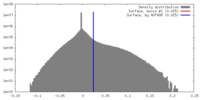
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : The cap portion of an extracellular contractile injection system ...
| Entire | Name: The cap portion of an extracellular contractile injection system in marine bacterium Algoriphagus machipongonensis |
|---|---|
| Components |
|
-Supramolecule #1: The cap portion of an extracellular contractile injection system ...
| Supramolecule | Name: The cap portion of an extracellular contractile injection system in marine bacterium Algoriphagus machipongonensis type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Algoriphagus machipongonensis (bacteria) Algoriphagus machipongonensis (bacteria) |
-Macromolecule #1: cap protein (Algo1)
| Macromolecule | Name: cap protein (Algo1) / type: protein_or_peptide / ID: 1 / Details: cap protein (Algo1) / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Algoriphagus machipongonensis (bacteria) Algoriphagus machipongonensis (bacteria) |
| Molecular weight | Theoretical: 22.981086 KDa |
| Sequence | String: MIFEVLKILT DEVNQNFKGL EMEDSEVVLN NVALIDSQQD VATELQNKVI LSMINLREEV TMKNFPNNVL EGTKVTYKNP KLNINLFLI FCANRTGYKK SLSDLSRILE FFQHKSVFTQ SNTSFDRDLE EMENVKNFRF TMELFTPTFE ELNYIWGTLG G RQYPSVFY ...String: MIFEVLKILT DEVNQNFKGL EMEDSEVVLN NVALIDSQQD VATELQNKVI LSMINLREEV TMKNFPNNVL EGTKVTYKNP KLNINLFLI FCANRTGYKK SLSDLSRILE FFQHKSVFTQ SNTSFDRDLE EMENVKNFRF TMELFTPTFE ELNYIWGTLG G RQYPSVFY KLNLIVIDRD ATTSEEGVIT NIHRNYETL UniProtKB: Pvc16 N-terminal domain-containing protein |
-Macromolecule #2: cap adaptor protein (Algo2)
| Macromolecule | Name: cap adaptor protein (Algo2) / type: protein_or_peptide / ID: 2 / Details: cap adaptor protein (Algo2) / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Algoriphagus machipongonensis (bacteria) Algoriphagus machipongonensis (bacteria) |
| Molecular weight | Theoretical: 33.083371 KDa |
| Sequence | String: MQVSSSFRSF LKLDILHSYF LNDGEKDFSS MNEEESKTQL KSYNWKDFLE IYPSQKTSHM MRGNKIFFKS FNDSIILAIK VESGTENQP FNELYEDESM TFLLSLKDQY FGNYTDLDLA DQLLYFSNKT PVLPEAFTFK PIDRINQSGT VGEEYLYEGE N KKHLLEEA ...String: MQVSSSFRSF LKLDILHSYF LNDGEKDFSS MNEEESKTQL KSYNWKDFLE IYPSQKTSHM MRGNKIFFKS FNDSIILAIK VESGTENQP FNELYEDESM TFLLSLKDQY FGNYTDLDLA DQLLYFSNKT PVLPEAFTFK PIDRINQSGT VGEEYLYEGE N KKHLLEEA HLNPGGGVLG IIQIYMKGDT PVLSLINNDG TLKNSLPHFK IHFSNRKSTW KYINLKDDFE TETKKDYPLT KF GFILLDK KSDFISPPAH FEKYVFPNPD ARRIKITPTK NYSEIFI UniProtKB: Uncharacterized protein |
-Macromolecule #3: Phage tail protein
| Macromolecule | Name: Phage tail protein / type: protein_or_peptide / ID: 3 / Number of copies: 12 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Algoriphagus machipongonensis (bacteria) Algoriphagus machipongonensis (bacteria) |
| Molecular weight | Theoretical: 16.375458 KDa |
| Sequence | String: MSYPLSKFHF SVEWGGTKIG FTEVSGLDLE TEIIEYRHGA SPEYSKIKMP GMQKFSNITL KRGTFKSDNE YFQWYNTINL NKVERRDLT ISLLNEEHEP VVTWKVKNAW PLKVQSTDLK GDGNEVAIES MELAHEGLVI QNE UniProtKB: Phage tail protein |
-Macromolecule #4: Putative phage tail sheath protein FI
| Macromolecule | Name: Putative phage tail sheath protein FI / type: protein_or_peptide / ID: 4 Details: the residues (288-320aa) are not built up in the model and the residues (430-446aa) are assigned as poly-alanine due to the poor density map. Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Algoriphagus machipongonensis (bacteria) Algoriphagus machipongonensis (bacteria) |
| Molecular weight | Theoretical: 76.319039 KDa |
| Sequence | String: MATYKTPGVY IEEITKFPPS VAQVETAIPA FIGYTQFART KPSVDSDDLI LKPKRISSLL DFTTYYGGAQ NEQGITVKLT DTLIEGAEN RTINVPEPTF KSPYLMFYSL QMYFANGGGP CYIVSTGVYD DWSDSETPPT INFSDLESGL AVIRKEDEPT L LLFPDATN ...String: MATYKTPGVY IEEITKFPPS VAQVETAIPA FIGYTQFART KPSVDSDDLI LKPKRISSLL DFTTYYGGAQ NEQGITVKLT DTLIEGAEN RTINVPEPTF KSPYLMFYSL QMYFANGGGP CYIVSTGVYD DWSDSETPPT INFSDLESGL AVIRKEDEPT L LLFPDATN LPTDDEFYSL YNSALMQCND LQDRFTILDT YSDQTYNDGV EDLDPIPALR NGINLTKDYL KYGAAYYPFV QT ILNYQYS ADEIVIQHLS YNPNAIATAL DNLNAVNGPT FIDAILDDLR DLSLPDISGE ISDAVGFMYD DVDGFDIDGT FTT NSVKVA NFASLVESVL STLNELIDAK EEINKDVNSA IASSEEDNAI KTAISDALDV FNEDFEGADK IESVAKNLSD LLIK IKQAD TNTKVENVLS INALNFSAEF EKLLTYDVNT GLTASVTLDL FANIGTRLDD IIAAVSAAEP IDVNNGKLNG RLLSD IEPL DNATYNTILL EINSHKVTLP PSSSMAGAYA RVDNDRGVWK SPANIGLNYV SKPSVTVSHE EQESMNVHGT GKSVNA IRS FVGKGTLVWG ARTLAGNDNE WRYISVRRFF NMAEESIKKA TEQFVFEPND GNTWVRVRAM IENFLILQWR AGALAGA KP EHAFYVKVGL GQTMTAQDIL EGNMNVEIGL AVVRPAEFII LKFSHKMQES UniProtKB: Putative phage tail sheath protein FI |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Grid | Model: Quantifoil / Material: GOLD / Mesh: 200 |
| Vitrification | Cryogen name: ETHANE-PROPANE / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: EMDB MAP EMDB ID: |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 2.5 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 65059 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL |
|---|---|
| Output model |  PDB-7adz: |
 Movie
Movie Controller
Controller



























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)