[English] 日本語
 Yorodumi
Yorodumi- EMDB-12034: Cryo-EM structure of the contractile injection system cap complex... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-12034 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the contractile injection system cap complex from Anabaena PCC7120 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | contractile tail / injection system / macromolecular machine / contractile protein / PROTEIN TRANSPORT | |||||||||
| Function / homology |  Function and homology information Function and homology information | |||||||||
| Biological species |  Nostoc sp. PCC 7120 = FACHB-418 (bacteria) / Nostoc sp. PCC 7120 = FACHB-418 (bacteria) /  Nostoc sp. (strain PCC 7120 / SAG 25.82 / UTEX 2576) (bacteria) Nostoc sp. (strain PCC 7120 / SAG 25.82 / UTEX 2576) (bacteria) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.8 Å | |||||||||
 Authors Authors | Eisenstein F / Weiss GL | |||||||||
 Citation Citation |  Journal: Nat Microbiol / Year: 2022 Journal: Nat Microbiol / Year: 2022Title: Structure of a thylakoid-anchored contractile injection system in multicellular cyanobacteria. Authors: Gregor L Weiss / Fabian Eisenstein / Ann-Katrin Kieninger / Jingwei Xu / Hannah A Minas / Milena Gerber / Miki Feldmüller / Iris Maldener / Karl Forchhammer / Martin Pilhofer /    Abstract: Contractile injection systems (CISs) mediate cell-cell interactions by phage tail-like structures, using two distinct modes of action: extracellular CISs are released into the medium, while type 6 ...Contractile injection systems (CISs) mediate cell-cell interactions by phage tail-like structures, using two distinct modes of action: extracellular CISs are released into the medium, while type 6 secretion systems (T6SSs) are attached to the cytoplasmic membrane and function upon cell-cell contact. Here, we characterized a CIS in the multicellular cyanobacterium Anabaena, with features distinct from extracellular CISs and T6SSs. Cryo-electron tomography of focused ion beam-milled cells revealed that CISs were anchored in thylakoid membrane stacks, facing the cell periphery. Single particle cryo-electron microscopy showed that this unique in situ localization was mediated by extensions of tail fibre and baseplate components. On stress, cyanobacteria induced the formation of ghost cells, presenting thylakoid-anchored CISs to the environment. Functional assays suggest that these CISs may mediate ghost cell formation and/or interactions of ghost cells with other organisms. Collectively, these data provide a framework for understanding the evolutionary re-engineering of CISs and potential roles of these CISs in cyanobacterial programmed cell death. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_12034.map.gz emd_12034.map.gz | 479.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-12034-v30.xml emd-12034-v30.xml emd-12034.xml emd-12034.xml | 20.7 KB 20.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_12034_fsc.xml emd_12034_fsc.xml | 18 KB | Display |  FSC data file FSC data file |
| Images |  emd_12034.png emd_12034.png | 213.5 KB | ||
| Filedesc metadata |  emd-12034.cif.gz emd-12034.cif.gz | 6.2 KB | ||
| Others |  emd_12034_half_map_1.map.gz emd_12034_half_map_1.map.gz emd_12034_half_map_2.map.gz emd_12034_half_map_2.map.gz | 407.8 MB 407.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-12034 http://ftp.pdbj.org/pub/emdb/structures/EMD-12034 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12034 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12034 | HTTPS FTP |
-Related structure data
| Related structure data |  7b5iMC  7b5hC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_12034.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_12034.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.1 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
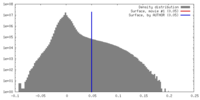
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Half map: #1
| File | emd_12034_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
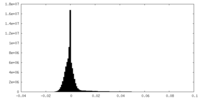
| Density Histograms |
-Half map: #2
| File | emd_12034_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
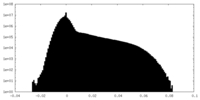
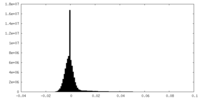
| Density Histograms |
- Sample components
Sample components
-Entire : Cap complex of the Anabaena PCC7120 contractile injection system
| Entire | Name: Cap complex of the Anabaena PCC7120 contractile injection system |
|---|---|
| Components |
|
-Supramolecule #1: Cap complex of the Anabaena PCC7120 contractile injection system
| Supramolecule | Name: Cap complex of the Anabaena PCC7120 contractile injection system type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Nostoc sp. PCC 7120 = FACHB-418 (bacteria) Nostoc sp. PCC 7120 = FACHB-418 (bacteria) |
-Macromolecule #1: All3327 protein
| Macromolecule | Name: All3327 protein / type: protein_or_peptide / ID: 1 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Nostoc sp. (strain PCC 7120 / SAG 25.82 / UTEX 2576) (bacteria) Nostoc sp. (strain PCC 7120 / SAG 25.82 / UTEX 2576) (bacteria)Strain: PCC 7120 / SAG 25.82 / UTEX 2576 |
| Molecular weight | Theoretical: 21.935064 KDa |
| Sequence | String: MISDAMRLIQ VALQRYILEF EPELGLSQVV IIENIAMAEE LGGQNNQING HVVMSLVNLQ EETTLKNSPH YRLDNGRTIY QNPPVNLNL FILFSALHNQ YETSLRLLSR VVEFFQWQKE LSFTTTPGIG GISRDLRILP DLYSLTFEQL NHLWGALGGK Q VPFVLYRA ...String: MISDAMRLIQ VALQRYILEF EPELGLSQVV IIENIAMAEE LGGQNNQING HVVMSLVNLQ EETTLKNSPH YRLDNGRTIY QNPPVNLNL FILFSALHNQ YETSLRLLSR VVEFFQWQKE LSFTTTPGIG GISRDLRILP DLYSLTFEQL NHLWGALGGK Q VPFVLYRA RILSLEAPKR QAEGSTITEI YINE UniProtKB: All3327 protein |
-Macromolecule #2: All3326 protein
| Macromolecule | Name: All3326 protein / type: protein_or_peptide / ID: 2 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Nostoc sp. (strain PCC 7120 / SAG 25.82 / UTEX 2576) (bacteria) Nostoc sp. (strain PCC 7120 / SAG 25.82 / UTEX 2576) (bacteria)Strain: PCC 7120 / SAG 25.82 / UTEX 2576 |
| Molecular weight | Theoretical: 45.348633 KDa |
| Sequence | String: MKILYKKILN LELWHDFYLG QPNTPGSLPN NYDISRTLAL VPTQECLRVL ANLRWVFRPQ LYGASLFANV NAAPSGQFPT IFPIDRVYR LTFWLVVSDR YFANFTNLSL INSRNQIYYF SNLSGNEGHA LFLTQPLSAY TTNNEYQLGQ LVTHADKTLE S LTYQGNAT ...String: MKILYKKILN LELWHDFYLG QPNTPGSLPN NYDISRTLAL VPTQECLRVL ANLRWVFRPQ LYGASLFANV NAAPSGQFPT IFPIDRVYR LTFWLVVSDR YFANFTNLSL INSRNQIYYF SNLSGNEGHA LFLTQPLSAY TTNNEYQLGQ LVTHADKTLE S LTYQGNAT NIPNPSDWDS LPASQYVSEL DHLPRQGTYR TQVITNANPD NTYNFTLVNT NEQESWAIDV IVPDTHKSGE PF STSLNFV GQTPGHYRLL ENDTQVAEFV LVDNSLPEAF ALVEVILNPE LVPSAFSLLQ ASAGQTFIQP KTYVIRFKNR ATR WRYRYE QPHGCSAANL PSYFNLIDTH TYATARPIGL RQRPDSLLND CQDRPLPAPS ITLIQPETDG SQRIARIFSD IYL UniProtKB: All3326 protein |
-Macromolecule #3: All3325 protein
| Macromolecule | Name: All3325 protein / type: protein_or_peptide / ID: 3 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Nostoc sp. (strain PCC 7120 / SAG 25.82 / UTEX 2576) (bacteria) Nostoc sp. (strain PCC 7120 / SAG 25.82 / UTEX 2576) (bacteria)Strain: PCC 7120 / SAG 25.82 / UTEX 2576 |
| Molecular weight | Theoretical: 53.23616 KDa |
| Sequence | String: MPSTYKTPGV YIEEISKFPP SIAQVETAIP AFIGYTQIAK VGVENFHTDA DNLILRPVRI TSLLEYEQFF GKAINETTIQ VVIQDTTDS RGNLTERKAS ARITSPSPHN LYYSMQAYFA NGGGPCYIVS VGPMSNTGTI QLEALQNGLA EVAKEDEVTL L VFPESQSL ...String: MPSTYKTPGV YIEEISKFPP SIAQVETAIP AFIGYTQIAK VGVENFHTDA DNLILRPVRI TSLLEYEQFF GKAINETTIQ VVIQDTTDS RGNLTERKAS ARITSPSPHN LYYSMQAYFA NGGGPCYIVS VGPMSNTGTI QLEALQNGLA EVAKEDEVTL L VFPESQSL SDENYAALMS AALEQCANLQ DRFTVMDLKL PATRPIPANA IVGASNAFRD LSLPQDNLKY GACYAPDIET IF NYFYQED AVTIFRSVNG GAEEQDTLTM AGYNPANGGD GIQYALIESA IDQLPLILPP SPLVVGQYAR TDNTRGVWKA PAN VALSSV IKPVLKITNE QQNNLNVHPT GKSINAIRAF TGKGTLIWGA RTLAGNDNEW RYVSVRRFFN MAEESIKKGS EPFV FEPND ANTWTKVKAM IENFLTLQWR AGALAGAKPE QAFYVKIGLN ETMTALDILE GRMIVEIGMA VVRPAEFIIL KFSHK MQES UniProtKB: All3325 protein |
-Macromolecule #4: All3324 protein
| Macromolecule | Name: All3324 protein / type: protein_or_peptide / ID: 4 / Number of copies: 12 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Nostoc sp. (strain PCC 7120 / SAG 25.82 / UTEX 2576) (bacteria) Nostoc sp. (strain PCC 7120 / SAG 25.82 / UTEX 2576) (bacteria)Strain: PCC 7120 / SAG 25.82 / UTEX 2576 |
| Molecular weight | Theoretical: 16.459543 KDa |
| Sequence | String: MAEYPLPKFH FQVDWGGSRL GFTEVSGLDV ETEVIEYREG NLPQYHKLKM PGMQKFSNIT MKRGTFQGDN DFYKWWNTVA LNTIERRDL TISLLNEKHE PVVVWKVNRA WPTKVQSTDL KGDGNEVAIE SIEVAHEGLT IQNG UniProtKB: All3324 protein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Grid | Model: Quantifoil / Material: COPPER / Support film - Material: CARBON / Support film - topology: CONTINUOUS |
| Vitrification | Cryogen name: ETHANE-PROPANE / Chamber humidity: 100 % / Chamber temperature: 281 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Detector mode: COUNTING / Number grids imaged: 4 / Number real images: 19000 / Average electron dose: 52.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Calibrated defocus max: 1.5 µm / Calibrated defocus min: 0.9 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)