-Search query
-Search result
Showing all 29 items for (author: doki & c)

EMDB-53046: 
Mouse otoferlin (216-1931) in complex with an MSP2N2 lipid nanodisc (30 mol% DOPS, 10 mol% PI(4,5)P2)
Method: single particle / : Cretu C, Moser T

PDB-9qe2: 
Mouse otoferlin (216-1931) in complex with an MSP2N2 lipid nanodisc (30 mol% DOPS, 10 mol% PI(4,5)P2)
Method: single particle / : Cretu C, Moser T

EMDB-54802: 
Mouse otoferlin (216-1931) in the lipid-free, Ca2+-bound state, "open" conformation (class 2)
Method: single particle / : Cretu C, Moser T

EMDB-54805: 
Mouse otoferlin (216-1931) in complex with a lipid nanodisc (comprising 25% PS and 5% PIP2)
Method: single particle / : Cretu C, Moser T

EMDB-54809: 
Mouse otoferlin (216-1931) in the lipid-free Ca2+-bound state, "open" conformation (class 1)
Method: single particle / : Cretu C, Moser T

EMDB-54827: 
Mouse otoferlin (residues 216-1931) in the lipid-bound state (merged datasets)
Method: single particle / : Cretu C, Moser T

EMDB-54883: 
Mouse otoferlin (216-1931) in the lipid-free, Ca2+-free state ("loose" conformation)
Method: single particle / : Cretu C, Moser T

EMDB-54923: 
Mouse otoferlin (216-1931) in the lipid-free Ca2+-bound state, "closed-like" conformation
Method: single particle / : Cretu C, Moser T

PDB-9se5: 
Mouse otoferlin (216-1931) in the lipid-free, Ca2+-bound state, "open" conformation (class 2)
Method: single particle / : Cretu C, Moser T

PDB-9sea: 
Mouse otoferlin (216-1931) in complex with a lipid nanodisc (comprising 25% PS and 5% PIP2)
Method: single particle / : Cretu C, Moser T

PDB-9seg: 
Mouse otoferlin (216-1931) in the lipid-free Ca2+-bound state, "open" conformation (class 1)
Method: single particle / : Cretu C, Moser T

PDB-9sfl: 
Mouse otoferlin (residues 216-1931) in the lipid-bound state (merged datasets)
Method: single particle / : Cretu C, Moser T

PDB-9sh0: 
Mouse otoferlin (216-1931) in the lipid-free, Ca2+-free state ("loose" conformation)
Method: single particle / : Cretu C, Moser T

PDB-9si1: 
Mouse otoferlin (216-1931) in the lipid-free Ca2+-bound state, "closed-like" conformation
Method: single particle / : Cretu C, Moser T

EMDB-16813: 
Tomogram of GBP1 coatomers assembled on brain polar lipid-derived small unilamellar vesicles.
Method: electron tomography / : Kuhm TI, Jakobi AJ
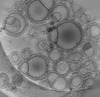
EMDB-16814: 
Tomogram of GBP1 coatomers assembled on brain polar lipid-derived small unilamellar vesicles.
Method: electron tomography / : Kuhm TI, Jakobi AJ
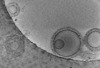
EMDB-16815: 
Tomogram of GBP1 coatomers assembled on brain polar lipid-derived small unilamellar vesicles.
Method: electron tomography / : Kuhm TI, Jakobi AJ

EMDB-16794: 
Cryo-EM structure of the human GBP1 dimer bound to GDP-AlF3
Method: single particle / : Kuhm TI, Jakobi AJ

PDB-8cqb: 
Cryo-EM structure of the human GBP1 dimer bound to GDP-AlF3
Method: single particle / : Kuhm TI, Jakobi AJ

EMDB-15646: 
Wild type hexamer oxalyl-CoA synthetase (OCS)
Method: single particle / : Lill P, Burgi J, Raunser S, Wilmanns M, Gatsogiannis C

PDB-8atd: 
Wild type hexamer oxalyl-CoA synthetase (OCS)
Method: single particle / : Lill P, Burgi J, Raunser S, Wilmanns M, Gatsogiannis C

EMDB-26357: 
CryoEM structure of the Candida albicans Aro1 dimer
Method: single particle / : Quade B, Borek D, Otwinowski Z

EMDB-26358: 
Structure of DHQS/EPSPS dimer from Candida albicans Aro1
Method: single particle / : Quade B, Borek D, Otwinowski Z

EMDB-26359: 
Structure of the SK/DHQase/DHSD dimer from Candida albicans Aro1
Method: single particle / : Quade B, Borek D, Otwinowski Z

PDB-7u5s: 
CryoEM structure of the Candida albicans Aro1 dimer
Method: single particle / : Quade B, Borek D, Otwinowski Z, Center for Structural Genomics of Infectious Diseases (CSGID)

PDB-7u5t: 
Structure of DHQS/EPSPS dimer from Candida albicans Aro1
Method: single particle / : Quade B, Borek D, Otwinowski Z, Center for Structural Genomics of Infectious Diseases (CSGID)

PDB-7u5u: 
Structure of the SK/DHQase/DHSD dimer from Candida albicans Aro1
Method: single particle / : Quade B, Borek D, Otwinowski Z, Center for Structural Genomics of Infectious Diseases (CSGID)

EMDB-9637: 
5R-MAP4, kinesin-1, and microtubule complex
Method: helical / : Shigematsu H, Imasaki T, Doki C, Sumi T, Aoki M, Uchikubo-Kamo T, Sakamoto A, Tokuraku K, Shirouzu M, Nitta R

EMDB-9638: 
4R-MAP4, kinesin-1, and microtubule complex
Method: helical / : Shigematsu H, Imasaki T, Doki C, Sumi T, Aoki M, Uchikubo-Kamo T, Sakamoto A, Tokuraku K, Shirouzu M, Nitta R
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
