+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Wild type hexamer oxalyl-CoA synthetase (OCS) | ||||||||||||||||||
 Map data Map data | |||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||
 Keywords Keywords | Peroxisome / Oxalyl-CoA ligase / Oligomer / Yeast / LIGASE | ||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationoxalate-CoA ligase / oxalate-CoA ligase activity / oxalate catabolic process / medium-chain fatty acid-CoA ligase activity / peroxisomal membrane / peroxisomal matrix / fatty acid metabolic process / mRNA binding / ATP binding / cytoplasm Similarity search - Function | ||||||||||||||||||
| Biological species |  | ||||||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.1 Å | ||||||||||||||||||
 Authors Authors | Lill P / Burgi J / Raunser S / Wilmanns M / Gatsogiannis C | ||||||||||||||||||
| Funding support |  Germany, 5 items Germany, 5 items
| ||||||||||||||||||
 Citation Citation |  Journal: Biol Chem / Year: 2023 Journal: Biol Chem / Year: 2023Title: Asymmetric horseshoe-like assembly of peroxisomal yeast oxalyl-CoA synthetase. Authors: Jérôme Bürgi / Pascal Lill / Evdokia-Anastasia Giannopoulou / Cy M Jeffries / Grzegorz Chojnowski / Stefan Raunser / Christos Gatsogiannis / Matthias Wilmanns /  Abstract: Oxalyl-CoA synthetase from is one of the most abundant peroxisomal proteins in yeast and hence has become a model to study peroxisomal translocation. It contains a C-terminal Peroxisome Targeting ...Oxalyl-CoA synthetase from is one of the most abundant peroxisomal proteins in yeast and hence has become a model to study peroxisomal translocation. It contains a C-terminal Peroxisome Targeting Signal 1, which however is partly dispensable, suggesting additional receptor bindings sites. To unravel any additional features that may contribute to its capacity to be recognized as peroxisomal target, we determined its assembly and overall architecture by an integrated structural biology approach, including X-ray crystallography, single particle cryo-electron microscopy and small angle X-ray scattering. Surprisingly, it assembles into mixture of concentration-dependent dimers, tetramers and hexamers by dimer self-association. Hexameric particles form an unprecedented asymmetric horseshoe-like arrangement, which considerably differs from symmetric hexameric assembly found in many other protein structures. A single mutation within the self-association interface is sufficient to abolish any higher-level oligomerization, resulting in a homogenous dimeric assembly. The small C-terminal domain of yeast Oxalyl-CoA synthetase is connected by a partly flexible hinge with the large N-terminal domain, which provides the sole basis for oligomeric assembly. Our data provide a basis to mechanistically study peroxisomal translocation of this target. | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_15646.map.gz emd_15646.map.gz | 4.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-15646-v30.xml emd-15646-v30.xml emd-15646.xml emd-15646.xml | 23.7 KB 23.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_15646.png emd_15646.png | 39.7 KB | ||
| Masks |  emd_15646_msk_1.map emd_15646_msk_1.map | 23 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-15646.cif.gz emd-15646.cif.gz | 6.7 KB | ||
| Others |  emd_15646_additional_1.map.gz emd_15646_additional_1.map.gz emd_15646_additional_2.map.gz emd_15646_additional_2.map.gz emd_15646_half_map_1.map.gz emd_15646_half_map_1.map.gz emd_15646_half_map_2.map.gz emd_15646_half_map_2.map.gz | 1.7 MB 10.3 MB 12.1 MB 11.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-15646 http://ftp.pdbj.org/pub/emdb/structures/EMD-15646 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15646 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15646 | HTTPS FTP |
-Related structure data
| Related structure data |  8atdMC  8affC  8afgC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_15646.map.gz / Format: CCP4 / Size: 23 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_15646.map.gz / Format: CCP4 / Size: 23 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.09 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_15646_msk_1.map emd_15646_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
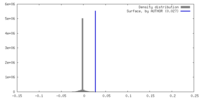
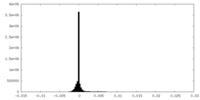
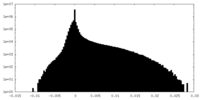
| Density Histograms |
-Additional map: #2
| File | emd_15646_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
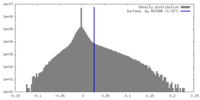
| Density Histograms |
-Additional map: #1
| File | emd_15646_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
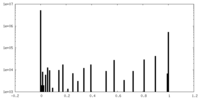
| Density Histograms |
-Half map: #2
| File | emd_15646_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
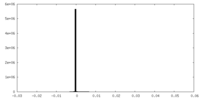
| Density Histograms |
-Half map: #1
| File | emd_15646_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Hexameric complex of the oxalyl-CoA synthetase Pcs60p
| Entire | Name: Hexameric complex of the oxalyl-CoA synthetase Pcs60p |
|---|---|
| Components |
|
-Supramolecule #1: Hexameric complex of the oxalyl-CoA synthetase Pcs60p
| Supramolecule | Name: Hexameric complex of the oxalyl-CoA synthetase Pcs60p / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 360 KDa |
-Macromolecule #1: Oxalate--CoA ligase
| Macromolecule | Name: Oxalate--CoA ligase / type: protein_or_peptide / ID: 1 / Number of copies: 6 / Enantiomer: LEVO / EC number: oxalate-CoA ligase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 48.135832 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: TVTASFNDTF SVSDNVAVIV PETDTQVTYR DLSHMVGHFQ TMFTNPNSPL YGAVFRQDTV AISMRNGLEF IVAFLGATMD AKIGAPLNP NYKEKEFNFY LNDLKSKAIC VPKGTTKLQS SEILKSASTF GCFIVELAFD ATRFRVEYDI YSPEDNYKRV I YRSLNNAK ...String: TVTASFNDTF SVSDNVAVIV PETDTQVTYR DLSHMVGHFQ TMFTNPNSPL YGAVFRQDTV AISMRNGLEF IVAFLGATMD AKIGAPLNP NYKEKEFNFY LNDLKSKAIC VPKGTTKLQS SEILKSASTF GCFIVELAFD ATRFRVEYDI YSPEDNYKRV I YRSLNNAK FVNTNPVKFP GFARSSDVAL ILHTSGTTST PKTVPLLHLN IVRSTLNIAN TYKLTPLDRS YVVMPLFHVH GL IGVLLST FRTQGSVVVP DGFHPKLFWD QFVKYNCNWF SCVPTISMIM LNMPKPNPFP HIRFIRSCSS ALAPATFHKL EKE FNAPVL EAYAMTEASH QMTSNNLPPG KRKPGTVGQP QGVTVVILDD NDNVLPPGKV GEVSIRGENV TLGYANNPKA NKEN FTKRE NYFRTGDQGY FDPEGFLVLT GRIKEL UniProtKB: Oxalate--CoA ligase |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.3 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| ||||||||||||
| Grid | Model: Quantifoil / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Chamber temperature: 298.15 K / Instrument: GATAN CRYOPLUNGE 3 Details: 90% humidity in the sample chamber. Gentle Blot function, 2.5 seconds. |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum |
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Detector mode: COUNTING / Digitization - Frames/image: 2-60 / Number grids imaged: 4 / Number real images: 5594 / Average electron dose: 63.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated defocus max: 2.2 µm / Calibrated defocus min: 0.7000000000000001 µm / Calibrated magnification: 105000 / Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Cs: 0.01 mm / Nominal defocus max: 2.2 µm / Nominal defocus min: 0.7000000000000001 µm / Nominal magnification: 105000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller



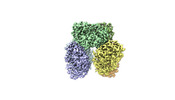

 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)