-Search query
-Search result
Showing 1 - 50 of 4,089 items for (author: cho & c)

EMDB entry, No image
EMDB-37593: 
Vibrio vulnificus MARTX effector duet (RDTND-RID) complexed with human Rac1 Q61L and calmodulin
EMDB-50358: 
In vitro-induced genome-releasing intermediate of Rhodobacter microvirus Ebor computed with C5 symmetry

EMDB-43475: 
Human GABAA receptor alpha1-beta2-gamma2 subtype in complex with GABA plus methaqualone

EMDB-43485: 
Human GABAA receptor alpha1-beta2-gamma2 subtype in complex with GABA plus PPTQ

PDB-8vqy: 
Human GABAA receptor alpha1-beta2-gamma2 subtype in complex with GABA plus methaqualone

PDB-8vrn: 
Human GABAA receptor alpha1-beta2-gamma2 subtype in complex with GABA plus PPTQ

EMDB-18881: 
AL amyloid fibril from the FOR010 light chain

EMDB-19818: 
AL amyloid fibril from the FOR103 light chain
EMDB-19395: 
CryoEM structure of recombinant human Bri2 BRICHOS oligomers

EMDB-19075: 
Conformational Landscape of the Type V-K CRISPR-associated TransposonIntegration Assembly CAST V-K composite map

EMDB-19282: 
Conformational Landscape of the Type V-K CRISPR-associated TransposonIntegration Assembly CAST V-K Cas12k domain local-refinement map

EMDB-19283: 
Conformational Landscape of the Type V-K CRISPR-associated TransposonIntegration Assembly CAST V-K TnsC domain local-refinement map

EMDB-19284: 
Conformational Landscape of the Type V-K CRISPR-associated TransposonIntegration Assembly CAST V-K TnsB domain local-refinement map

EMDB-19286: 
consensus map of the V-K CRISPR-associated Transposon Integration Assembly

PDB-8rdu: 
Conformational Landscape of the Type V-K CRISPR-associated TransposonIntegration Assembly CAST V-K composite map

PDB-8rkt: 
Conformational Landscape of the Type V-K CRISPR-associated TransposonIntegration Assembly CAST V-K Cas12k domain local-refinement map

PDB-8rku: 
Conformational Landscape of the Type V-K CRISPR-associated TransposonIntegration Assembly CAST V-K TnsC domain local-refinement map

PDB-8rkv: 
Conformational Landscape of the Type V-K CRISPR-associated TransposonIntegration Assembly CAST V-K TnsB domain local-refinement map

EMDB-39534: 
Cryo-EM structure of the human ABCB6 in complex with Cd(II):GSH
EMDB-39535: 
Cryo-EM structure of the human ABCB6 in complex with Cd(II):Phytochelatin 2

PDB-8yr3: 
Cryo-EM structure of the human ABCB6 in complex with Cd(II):GSH

PDB-8yr4: 
Cryo-EM structure of the human ABCB6 in complex with Cd(II):Phytochelatin 2

EMDB-38216: 
Cryo-EM structure of SARS-CoV-2 S-BQ.1 in complex with antibody O5C2

PDB-8xbf: 
Cryo-EM structure of SARS-CoV-2 S-BQ.1 in complex with antibody O5C2
EMDB-43712: 
Human EBP complexed with compound 1
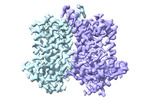
EMDB-43713: 
Human EBP complexed with compound 3a

PDB-8w0r: 
Human EBP complexed with compound 1

PDB-8w0s: 
Human EBP complexed with compound 3a

EMDB-36730: 
SARS-CoV-2 Spike RBD (dimer) in complex with two 2S-1244 nanobodies

EMDB-36735: 
Dimer of SARS-CoV-2 BA.2 spike and IBT-CoV144(C3 symmetry)

EMDB-36740: 
Dimer of SARS-CoV-2 BA.2 spike and IBT-CoV144(C1 symmetry)

PDB-8jys: 
SARS-CoV-2 Spike RBD (dimer) in complex with two 2S-1244 nanobodies

EMDB-50356: 
Empty capsid of Rhodobacter microvirus Ebor computed with I4 symmetry

EMDB-50357: 
Native capsid of Rhodobacter microvirus Ebor computed with I4 symmetry
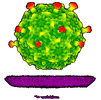
EMDB-50359: 
Rhodobacter microvirus Ebor attached to B10 host cell reconstructed by single particle analysis with applied C5 symmetry
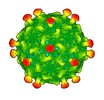
EMDB-50360: 
Rhodobacter microvirus Ebor attached to the outer membrane vesicle

EMDB-50361: 
Rhodobacter microvirus Ebor attached to the host cell reconstructed by subtomogram averaging

PDB-9ffg: 
Empty capsid of Rhodobacter microvirus Ebor computed with I4 symmetry

PDB-9ffh: 
Native capsid of Rhodobacter microvirus Ebor computed with I4 symmetry

EMDB-42074: 
Representative tomogram of Enterococcus faecium WT Com15

EMDB-42086: 
Representative tomogram of Enterococcus faecium SagA complementation strain

EMDB-42087: 
Representative tomogram of Enterococcus faecium SagA deletion strain

EMDB-18290: 
Cryo-EM structure of Cx26 gap junction K125E mutant in bicarbonate buffer (classification on hemichannel)

EMDB-18291: 
Cryo-EM structure of Cx26 solubilised in LMNG - hemichannel classification - NConst conformation

EMDB-18292: 
Cryo-EM structure of Cx26 solubilised in LMNG - Hemichannel classification NFlex conformation
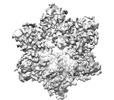
EMDB-18293: 
Cryo-EM structure of Cx26 solubilised in LMNG: classification on subunit A; Nconst-mon conformation

EMDB-18294: 
Cryo-EM structure of Cx26 solubilised in LMNG: classification on subunit A; NFlex conformation

EMDB-18295: 
Cryo-EM reconstruction of Cx26 gap junction K125R mutant (D6 symmetry)
EMDB-18296: 
Cryo-EM reconstruction of Cx26 gap junction K125E mutant in HEPES buffer

EMDB-18297: 
Cryo-EM reconstruction of Cx26 gap junction WT in HEPES buffer
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
