[English] 日本語
 Yorodumi
Yorodumi- EMDB-19282: Conformational Landscape of the Type V-K CRISPR-associated Transp... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Conformational Landscape of the Type V-K CRISPR-associated TransposonIntegration Assembly CAST V-K Cas12k domain local-refinement map | |||||||||
 Map data Map data | CAST V-K Cas12k domain local-refinement map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | CRISPR-associated Transposon genome editing transposition / DNA BINDING PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationcytosolic small ribosomal subunit / rRNA binding / structural constituent of ribosome / translation Similarity search - Function | |||||||||
| Biological species |  Scytonema hofmannii (bacteria) Scytonema hofmannii (bacteria) | |||||||||
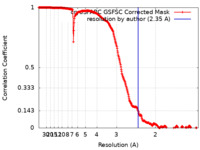
| Method | single particle reconstruction / cryo EM / Resolution: 2.35 Å | |||||||||
 Authors Authors | Tenjo-Castano F / Mesa P / Montoya G | |||||||||
| Funding support |  Denmark, 1 items Denmark, 1 items
| |||||||||
 Citation Citation |  Journal: Mol Cell / Year: 2024 Journal: Mol Cell / Year: 2024Title: Conformational landscape of the type V-K CRISPR-associated transposon integration assembly. Authors: Francisco Tenjo-Castaño / Nicholas Sofos / Luisa S Stutzke / Piero Temperini / Anders Fuglsang / Tillmann Pape / Pablo Mesa / Guillermo Montoya /  Abstract: CRISPR-associated transposons (CASTs) are mobile genetic elements that co-opt CRISPR-Cas systems for RNA-guided DNA transposition. CASTs integrate large DNA cargos into the attachment (att) site ...CRISPR-associated transposons (CASTs) are mobile genetic elements that co-opt CRISPR-Cas systems for RNA-guided DNA transposition. CASTs integrate large DNA cargos into the attachment (att) site independently of homology-directed repair and thus hold promise for eukaryotic genome engineering. However, the functional diversity and complexity of CASTs hinder an understanding of their mechanisms. Here, we present the high-resolution cryoelectron microscopy (cryo-EM) structure of the reconstituted ∼1 MDa post-transposition complex of the type V-K CAST, together with different assembly intermediates and diverse TnsC filament lengths, thus enabling the recapitulation of the integration complex formation. The results of mutagenesis experiments probing the roles of specific residues and TnsB-binding sites show that transposition activity can be enhanced and suggest that the distance between the PAM and att sites is determined by the lengths of the TnsB C terminus and the TnsC filament. This singular model of RNA-guided transposition provides a foundation for repurposing the system for genome-editing applications. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_19282.map.gz emd_19282.map.gz | 98.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-19282-v30.xml emd-19282-v30.xml emd-19282.xml emd-19282.xml | 22.2 KB 22.2 KB | Display Display |  EMDB header EMDB header |
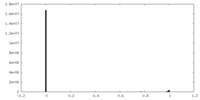
| FSC (resolution estimation) |  emd_19282_fsc.xml emd_19282_fsc.xml | 28.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_19282.png emd_19282.png | 182.6 KB | ||
| Masks |  emd_19282_msk_1.map emd_19282_msk_1.map | 106.1 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-19282.cif.gz emd-19282.cif.gz | 7.2 KB | ||
| Others |  emd_19282_half_map_1.map.gz emd_19282_half_map_1.map.gz emd_19282_half_map_2.map.gz emd_19282_half_map_2.map.gz | 98.6 MB 98.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-19282 http://ftp.pdbj.org/pub/emdb/structures/EMD-19282 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-19282 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-19282 | HTTPS FTP |
-Related structure data
| Related structure data |  8rktMC  8axaC  8axbC  8rduC  8rkuC  8rkvC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_19282.map.gz / Format: CCP4 / Size: 106.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_19282.map.gz / Format: CCP4 / Size: 106.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | CAST V-K Cas12k domain local-refinement map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.728 Å | ||||||||||||||||||||||||||||||||||||
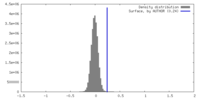
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_19282_msk_1.map emd_19282_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
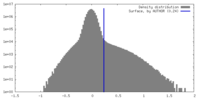
| Density Histograms |
-Half map: CAST V-K Cas12k domain local refinement, half-A map
| File | emd_19282_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | CAST V-K Cas12k domain local refinement, half-A map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: CAST V-K Cas12k domain local refinement, half-B map
| File | emd_19282_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | CAST V-K Cas12k domain local refinement, half-B map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
+Entire : recognition complex
+Supramolecule #1: recognition complex
+Macromolecule #1: sgRNA
+Macromolecule #2: Non-target strand - LE
+Macromolecule #3: Target strand - LE
+Macromolecule #4: ShCas12k
+Macromolecule #5: Small ribosomal subunit protein uS15
+Macromolecule #6: TniQ
+Macromolecule #7: MAGNESIUM ION
+Macromolecule #8: ZINC ION
+Macromolecule #9: water
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7 |
|---|---|
| Grid | Model: UltrAuFoil R0./1 / Material: GOLD / Mesh: 300 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Number real images: 19936 / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.8 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Protocol: AB INITIO MODEL |
|---|---|
| Output model |  PDB-8rkt: |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)