+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | AL amyloid fibril from the FOR103 light chain | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Amyloid fibril / AL amyloid / PROTEIN FIBRIL | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
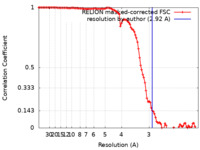
| Method | helical reconstruction / cryo EM / Resolution: 2.92 Å | |||||||||
 Authors Authors | Pfeiffer PB / Karimi-Farsijani S / Kupfer N / Schmidt M / Faendrich M | |||||||||
| Funding support |  Germany, 1 items Germany, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: Light chain mutations contribute to defining the fibril morphology in systemic AL amyloidosis. Authors: Sara Karimi-Farsijani / Peter Benedikt Pfeiffer / Sambhasan Banerjee / Julian Baur / Lukas Kuhn / Niklas Kupfer / Ute Hegenbart / Stefan O Schönland / Sebastian Wiese / Christian Haupt / ...Authors: Sara Karimi-Farsijani / Peter Benedikt Pfeiffer / Sambhasan Banerjee / Julian Baur / Lukas Kuhn / Niklas Kupfer / Ute Hegenbart / Stefan O Schönland / Sebastian Wiese / Christian Haupt / Matthias Schmidt / Marcus Fändrich /  Abstract: Systemic AL amyloidosis is one of the most frequently diagnosed forms of systemic amyloidosis. It arises from mutational changes in immunoglobulin light chains. To explore whether these mutations may ...Systemic AL amyloidosis is one of the most frequently diagnosed forms of systemic amyloidosis. It arises from mutational changes in immunoglobulin light chains. To explore whether these mutations may affect the structure of the formed fibrils, we determine and compare the fibril structures from several patients with cardiac AL amyloidosis. All patients are affected by light chains that contain an IGLV3-19 gene segment, and the deposited fibrils differ by the mutations within this common germ line background. Using cryo-electron microscopy, we here find different fibril structures in each patient. These data establish that the mutations of amyloidogenic light chains contribute to defining the fibril architecture and hence the structure of the pathogenic agent. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_19818.map.gz emd_19818.map.gz | 7.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-19818-v30.xml emd-19818-v30.xml emd-19818.xml emd-19818.xml | 15.7 KB 15.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_19818_fsc.xml emd_19818_fsc.xml | 10.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_19818.png emd_19818.png | 96 KB | ||
| Masks |  emd_19818_msk_1.map emd_19818_msk_1.map | 103 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-19818.cif.gz emd-19818.cif.gz | 5.3 KB | ||
| Others |  emd_19818_half_map_1.map.gz emd_19818_half_map_1.map.gz emd_19818_half_map_2.map.gz emd_19818_half_map_2.map.gz | 80.3 MB 80.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-19818 http://ftp.pdbj.org/pub/emdb/structures/EMD-19818 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-19818 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-19818 | HTTPS FTP |
-Validation report
| Summary document |  emd_19818_validation.pdf.gz emd_19818_validation.pdf.gz | 712.4 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_19818_full_validation.pdf.gz emd_19818_full_validation.pdf.gz | 711.9 KB | Display | |
| Data in XML |  emd_19818_validation.xml.gz emd_19818_validation.xml.gz | 17.9 KB | Display | |
| Data in CIF |  emd_19818_validation.cif.gz emd_19818_validation.cif.gz | 23.6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-19818 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-19818 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-19818 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-19818 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_19818.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_19818.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.04 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_19818_msk_1.map emd_19818_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_19818_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_19818_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : lambda 3 immunoglobulin light chain fragment
| Entire | Name: lambda 3 immunoglobulin light chain fragment |
|---|---|
| Components |
|
-Supramolecule #1: lambda 3 immunoglobulin light chain fragment
| Supramolecule | Name: lambda 3 immunoglobulin light chain fragment / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all / Details: residues 2-116 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: lambda 3 immunoglobulin light chain fragment, residues 2-116
| Macromolecule | Name: lambda 3 immunoglobulin light chain fragment, residues 2-116 type: protein_or_peptide / ID: 1 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 12.202262 KDa |
| Sequence | String: SELTQDPAVS VALGQTVRIT CQGDSLRSYY ASWYQQKSGQ APVLVIYSYN NRPSGIPDRF SGSNSGNTAS LTITGAQAED EADYYCNSR DSSGHHLVFG GGTKLTVLGQ PKAAPS |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | helical array |
- Sample preparation
Sample preparation
| Buffer | pH: 7 / Details: water |
|---|---|
| Grid | Model: C-flat-1.2/1.3 / Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 40 sec. |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Instrument: LEICA PLUNGER |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Detector mode: COUNTING / Number grids imaged: 1 / Number real images: 2788 / Average exposure time: 10.0 sec. / Average electron dose: 53.7 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.2 µm / Nominal magnification: 130000 |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: BACKBONE TRACE / Target criteria: correlation coefficient |
|---|---|
| Output model |  PDB-9eme: |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)