-Search query
-Search result
Showing 1 - 50 of 162 items for (author: barnes & g)

EMDB-42044: 
H2HA A/Japan/305/1957 complexed with mouse polyclonal Fab - wk 12, reoriented immunogen

EMDB-42045: 
H2HA A/Japan/305/1957 complexed with mouse polyclonal Fab - wk 12, control immunogen

EMDB-42046: 
H7N9 A/Shanghai/2/2013 complexed with mouse polyclonal Fab - wk 12, control immunogen

EMDB-42048: 
H3N2 A/Victoria/3/1975 complexed with mouse polyclonal Fab - wk 7, control immunogen

EMDB-42052: 
H3N2 A/Victoria/3/1975 complexed with mouse polyclonal Fab - wk 7, reoH2HA immunogen

EMDB-42056: 
H7N9 A/Shanghai/2/2013 complexed with mouse polyclonal Fab - wk 7, reoriented immunogen

EMDB-42058: 
H7N9 A/Shanghai/2/2013 complexed with mouse polyclonal Fab - wk 7, control immunogen

EMDB-42059: 
H7N9 A/Shanghai/2/2013 complexed with mouse polyclonal Fab - wk 7, reoriented immunogen

EMDB-29930: 
T. cruzi topoisomerase II alpha bound to dsDNA and the covalent inhibitor CT1

PDB-8gcc: 
T. cruzi topoisomerase II alpha bound to dsDNA and the covalent inhibitor CT1

EMDB-40094: 
17b10 fab in complex with full-length SARS-CoV-2 Spike G614 trimer

EMDB-40095: 
17B10 fab in complex with up-RBD of SARS-CoV-2 Spike G614 trimer

PDB-8gjm: 
17b10 fab in complex with full-length SARS-CoV-2 Spike G614 trimer

PDB-8gjn: 
17B10 fab in complex with up-RBD of SARS-CoV-2 Spike G614 trimer

EMDB-15971: 
SARS-CoV-2 Delta-RBD complexed with Fabs BA.2-36, BA.2-23, EY6A and COVOX-45

PDB-8bcz: 
SARS-CoV-2 Delta-RBD complexed with Fabs BA.2-36, BA.2-23, EY6A and COVOX-45

EMDB-27177: 
sd1.040 Fab in complex with SARS-CoV-2 Spike 2P glycoprotein

PDB-8d48: 
sd1.040 Fab in complex with SARS-CoV-2 Spike 2P glycoprotein

EMDB-26604: 
H1 Solomon Islands 2006 hemagglutinin in complex with Ab111
EMDB-26605: 
H1 Solomon Islands 2006 hemagglutinin in complex with Ab109

PDB-7umm: 
H1 Solomon Islands 2006 hemagglutinin in complex with Ab109

EMDB-26491: 
Cryo-EM structure of BG24 inferred germline Fabs with germline CDR3s and 10-1074 Fabs in complex with HIV-1 Env immunogen BG505-SOSIPv4.1-GT1 - Class 2

EMDB-26494: 
Cryo-EM map of BG24 Fabs with an inferred germline light chain and 10-1074 Fabs in complex with HIV-1 Env immunogen BG505-SOSIPv4.1-GT1 containing the N276 gp120 glycan- Class 2

EMDB-26495: 
Cryo-EM map of BG24 Fabs with an inferred germline light chain and 10-1074 Fabs in complex with HIV-1 Env immunogen BG505-SOSIPv4.1-GT1 containing the N276 gp120 glycan- Class 3
EMDB-26493: 
Cryo-EM structure of BG24 Fabs with an inferred germline light chain and 10-1074 Fabs in complex with HIV-1 Env immunogen BG505-SOSIPv4.1-GT1 containing the N276 gp120 glycan- Class 1

EMDB-26496: 
Cryo-EM structure of BG24 Fabs with an inferred germline CDRL1 and 10-1074 Fabs in complex with HIV-1 Env 6405-SOSIP.664

PDB-7ugp: 
Cryo-EM structure of BG24 Fabs with an inferred germline light chain and 10-1074 Fabs in complex with HIV-1 Env immunogen BG505-SOSIPv4.1-GT1 containing the N276 gp120 glycan- Class 1

PDB-7ugq: 
Cryo-EM structure of BG24 Fabs with an inferred germline CDRL1 and 10-1074 Fabs in complex with HIV-1 Env 6405-SOSIP.664
EMDB-26490: 
Cryo-EM structure of BG24 inferred germline Fabs with germline CDR3s and 10-1074 Fabs in complex with HIV-1 Env immunogen BG505-SOSIPv4.1-GT1 - Class 1
EMDB-26492: 
Cryo-EM structure of BG24 inferred germline Fabs with mature CDR3s and 10-1074 Fabs in complex with HIV-1 Env immunogen BG505-SOSIPv4.1-GT1

PDB-7ugn: 
Cryo-EM structure of BG24 inferred germline Fabs with germline CDR3s and 10-1074 Fabs in complex with HIV-1 Env immunogen BG505-SOSIPv4.1-GT1 - Class 1

PDB-7ugo: 
Cryo-EM structure of BG24 inferred germline Fabs with mature CDR3s and 10-1074 Fabs in complex with HIV-1 Env immunogen BG505-SOSIPv4.1-GT1
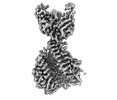
EMDB-27939: 
Structure of the human ACE2 receptor in complex with antibody Fab fragment, 05B04

EMDB-25039: 
Structure of H1 NC99 influenza hemagglutinin bound to Fab 310-63E6

EMDB-25040: 
Structure of H1 influenza hemagglutinin bound to Fab 310-39G10

PDB-7scn: 
Structure of H1 NC99 influenza hemagglutinin bound to Fab 310-63E6

PDB-7sco: 
Structure of H1 influenza hemagglutinin bound to Fab 310-39G10

EMDB-26443: 
Structure of the DU422 SOSIP.664 trimer in complex with neutralizing antibody Fab fragments 10-1074 and BG24

PDB-7ucg: 
Structure of the DU422 SOSIP.664 trimer in complex with neutralizing antibody Fab fragments 10-1074 and BG24
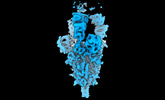
EMDB-14885: 
OMI-42 FAB IN COMPLEX WITH SARS-COV-2 BETA SPIKE GLYCOPROTEIN

EMDB-14886: 
OMI-38 FAB IN COMPLEX WITH SARS-COV-2 BETA SPIKE RBD (local refinement)

EMDB-14887: 
OMI-2 FAB IN COMPLEX WITH SARS-COV-2 BETA SPIKE GLYCOPROTEIN
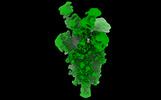
EMDB-14910: 
OMI-38 FAB IN COMPLEX WITH SARS-COV-2 BETA SPIKE

PDB-7zr7: 
OMI-42 FAB IN COMPLEX WITH SARS-COV-2 BETA SPIKE GLYCOPROTEIN

PDB-7zr8: 
OMI-38 FAB IN COMPLEX WITH SARS-COV-2 BETA SPIKE RBD (local refinement)

PDB-7zr9: 
OMI-2 FAB IN COMPLEX WITH SARS-COV-2 BETA SPIKE GLYCOPROTEIN

PDB-7zrc: 
OMI-38 FAB IN COMPLEX WITH SARS-COV-2 BETA SPIKE

EMDB-26429: 
Structure of the SARS-CoV-2 S 6P trimer in complex with the neutralizing antibody Fab fragment, C1520

EMDB-26430: 
Structure of the SARS-CoV-2 NTD in complex with C1520, local refinement

EMDB-26431: 
Structure of the SARS-CoV-2 S 6P trimer in complex with the neutralizing antibody Fab fragment, C1717
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
