-Search query
-Search result
Showing 1 - 50 of 234 items for (author: li & cp)

EMDB-35827: 
Structure of CbCas9 bound to 20-nucleotide complementary DNA substrate
Method: single particle / : Zhang S, Lin S, Liu JJG

EMDB-37652: 
Structure of CbCas9 bound to 6-nucleotide complementary DNA substrate
Method: single particle / : Zhang S, Lin S, Liu JJG

EMDB-37656: 
Structure of CbCas9-PcrIIC1 complex bound to 28-bp DNA substrate (20-nt complementary)
Method: single particle / : Zhang S, Lin S, Liu JJG

EMDB-37657: 
Structure of CbCas9-PcrIIC1 complex bound to 62-bp DNA substrate (symmetric 20-nt complementary)
Method: single particle / : Zhang S, Lin S, Liu JJG

EMDB-37762: 
Structure of CbCas9-PcrIIC1 complex bound to 62-bp DNA substrate (non-targeting complex)
Method: single particle / : Zhang S, Lin S, Liu JJG

EMDB-40815: 
BG505 SOSIP.664 in complex with wk26 human polyclonal antibodies C3V5, V1V3, N611 and base from participant 017
Method: single particle / : Karlinsey D, Ozorowski G, Ward AB

EMDB-40816: 
BG505 SOSIP.664 in complex with wk26 human polyclonal antibodies gp41-N611/FP and base from participant 03
Method: single particle / : Karlinsey D, Ozorowski G, Ward AB

EMDB-40817: 
BG505 SOSIP.664 in complex with wk26 human polyclonal antibodies gp41-N611/FP and base from participant 07
Method: single particle / : Karlinsey D, Ozorowski G, Ward AB

EMDB-40818: 
BG505 SOSIP.664 in complex with wk26 human polyclonal antibodies gp120-GH and base from participant 09
Method: single particle / : Karlinsey D, Ozorowski G, Ward AB

EMDB-40819: 
BG505 SOSIP.664 in complex with wk26 human polyclonal antibodies C3V5, V1V3, gp41-GH/FP and base from participant 11
Method: single particle / : Karlinsey D, Ozorowski G, Ward AB

EMDB-36776: 
The Anoxybacillus pushchinoensis ORF-less Group IIC Intron HYER1 at symmetric apo state
Method: single particle / : Zhu HZ, Liu JJG

EMDB-36777: 
The Anoxybacillus pushchinoensis ORF-less Group IIC Intron DR1 at symmetric pre-cleavage state
Method: single particle / : Zhu HZ, Liu JJG

EMDB-36778: 
The Anoxybacillus pushchinoensis ORF-less Group IIC Intron HYER1 at symmetric post cleavge state
Method: single particle / : Zhu HZ, Liu JJG

EMDB-36786: 
The Streptococcus azizii ORF-less Group IIC intron HYER2 at apo state
Method: single particle / : Zhu HZ, Liu JJG

EMDB-18889: 
Mouse teneurin-3 compact dimer - A1B1 isoform
Method: single particle / : Gogou C, Meijer DH

EMDB-18890: 
Mouse teneurin-3 non-compact subunit - A1B1 isoform
Method: single particle / : Gogou C, Meijer DH

EMDB-18891: 
Mouse teneurin-3 non-compact subunit - A0B0 isoform
Method: single particle / : Gogou C, Meijer DH

EMDB-19409: 
Mouse teneurin-3 compact dimer - A1B0 isoform
Method: single particle / : Gogou C, Meijer DH

EMDB-36779: 
The Anoxybacillus pushchinoensis ORF-less Group IIC Intron HYER1 with 10-nt TRS at symmetric apo state
Method: single particle / : Zhu HZ, Liu JJG

EMDB-18334: 
Cryo-EM structure of the inward-facing FLVCR1
Method: single particle / : Weng TH, Wu D, Safarian S

EMDB-18335: 
Cryo-EM structure of the inward-facing choline-bound FLVCR1
Method: single particle / : Weng TH, Wu D, Safarian S

EMDB-18336: 
Cryo-EM structure of the inward-facing FLVCR2
Method: single particle / : Weng TH, Wu D, Safarian S
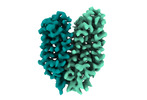
EMDB-18337: 
Cryo-EM structure of the outward-facing FLVCR2
Method: single particle / : Weng TH, Wu D, Safarian S

EMDB-18339: 
Cryo-EM structure of the inward-facing choline-bound FLVCR2
Method: single particle / : Weng TH, Wu D, Safarian S

EMDB-19009: 
Cryo-EM structure of the inward-facing ethanolamine-bound FLVCR1
Method: single particle / : Weng TH, Wu D, Safarian S

EMDB-40856: 
Single particle reconstruction of the human LINE-1 ORF2p without substrate (apo)
Method: single particle / : van Eeuwen T, Taylor MS, Rout MP
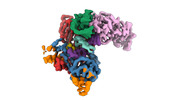
EMDB-40858: 
Structure of LINE-1 ORF2p with template:primer hybrid
Method: single particle / : van Eeuwen T, Taylor MS, Rout MP
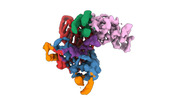
EMDB-40859: 
Structure of LINE-1 ORF2p with an oligo(A) template
Method: single particle / : van Eeuwen T, Taylor MS, Rout MP

PDB-8sxt: 
Structure of LINE-1 ORF2p with template:primer hybrid
Method: single particle / : van Eeuwen T, Taylor MS, Rout MP

PDB-8sxu: 
Structure of LINE-1 ORF2p with an oligo(A) template
Method: single particle / : van Eeuwen T, Taylor MS, Rout MP

EMDB-18900: 
Mouse teneurin-3 non-compact subunit - A0B1 isoform
Method: single particle / : Gogou C, Meijer DH

EMDB-18902: 
Mouse teneurin-3 non-compact subunit - A1B0 isoform
Method: single particle / : Gogou C, Meijer DH

EMDB-17804: 
Bat-Hp-CoV Nsp1 and eIF1 bound to the human 40S small ribosomal subunit
Method: single particle / : Schubert K, Karousis ED, Ban I, Lapointe CP, Leibundgut M, Baeumlin E, Kummerant E, Scaiola A, Schoenhut T, Ziegelmueller J, Puglisi JD, Muehlemann O, Ban N

EMDB-17805: 
MERS-CoV Nsp1 bound to the human 43S pre-initiation complex
Method: single particle / : Schubert K, Karousis ED, Ban I, Lapointe CP, Leibundgut M, Baeumlin E, Kummerant E, Scaiola A, Schoenhut T, Ziegelmueller J, Puglisi JD, Muehlemann O, Ban N

EMDB-29753: 
Tomogram of an apical end of a Toxoplasma Gondii tachyzoite displaying an extruded conoid
Method: electron tomography / : Martinez M, Chang YW, Mageswaran SK

EMDB-29754: 
Cryptosporidium parvum sporozoite apical end
Method: electron tomography / : Martinez M, Chang YW, Mageswaran SK
EMDB-29755: 
Cryptosporidium parvum sporozoite basal end
Method: electron tomography / : Martinez M, Chang YW, Mageswaran SK

EMDB-29784: 
Subtomogram average of the preconoidal rings from Cryptosporidium parvum sporozoites
Method: subtomogram averaging / : Martinez M, Mageswaran SK, Chang YW

EMDB-29791: 
Refined subtomogram average of the top preconoidal ring from Cryptosporidium parvum sporozoites
Method: subtomogram averaging / : Martinez M, Mageswaran SK, Chang YW

EMDB-29801: 
Refined subtomogram average of the bottom preconoidal ring from Cryptosporidium parvum sporozoites
Method: subtomogram averaging / : Martinez M, Mageswaran SK, Chang YW
EMDB-29808: 
Subtomogram average of the IMC surface filaments, top view, from Cryptosporidium parvum sporozoites
Method: subtomogram averaging / : Martinez M, Mageswaran SK, Chang YW

EMDB-29809: 
IMC surface filament, sawtooth conformation, from Cryptosporidium parvum sporozoites
Method: subtomogram averaging / : Martinez M, Mageswaran SK, Chang YW

EMDB-29810: 
IMC surface filament, side view, from Cryptosporidium parvum sporozoites
Method: subtomogram averaging / : Martinez M, Mageswaran SK, Chang YW

EMDB-29826: 
Upper preconoidal ring from Toxoplasma gondii tachyzoites
Method: subtomogram averaging / : Martinez M, Mageswaran SK, Chang YW

EMDB-29827: 
Lower preconoidal ring of Toxoplasma gondii tachyzoites
Method: subtomogram averaging / : Martinez M, Mageswaran SK, Chang YW

EMDB-29832: 
Preconoidal rings of Toxoplasma gondii tachyzoites
Method: subtomogram averaging / : Martinez M, Mageswaran SK, Chang YW

EMDB-29835: 
Basal IMC pore of Cryptosporidium parvum sporozoites
Method: subtomogram averaging / : Martinez M, Mageswaran SK, Chang YW

EMDB-29838: 
Preconoidal rings of Toxoplasma gondii tachyzoites with Formin1 conditionally depleted
Method: subtomogram averaging / : Martinez M, Mageswaran SK, Chang YW

EMDB-29839: 
Upper preconoidal ring of Toxoplasma gondii tachyzoites with Formin1 conditionally depleted
Method: subtomogram averaging / : Martinez M, Mageswaran SK, Chang YW

EMDB-29840: 
Lower preconoidal ring from Toxoplasma gondii tachyzoites with Formin1 conditionally depleted
Method: subtomogram averaging / : Martinez M, Mageswaran SK, Chang YW
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
