[English] 日本語
 Yorodumi
Yorodumi- EMDB-36779: The Anoxybacillus pushchinoensis ORF-less Group IIC Intron HYER1 ... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | The Anoxybacillus pushchinoensis ORF-less Group IIC Intron HYER1 with 10-nt TRS at symmetric apo state | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | ORF-less Group IIC intron / ribozymes / RNA | |||||||||
| Biological species |  Anoxybacillus pushchinoensis (bacteria) Anoxybacillus pushchinoensis (bacteria) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.41 Å | |||||||||
 Authors Authors | Zhu HZ / Liu JJG | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Science / Year: 2024 Journal: Science / Year: 2024Title: Hydrolytic endonucleolytic ribozyme (HYER) is programmable for sequence-specific DNA cleavage. Authors: Zi-Xian Liu / Shouyue Zhang / Han-Zhou Zhu / Zhi-Hang Chen / Yun Yang / Long-Qi Li / Yuan Lei / Yun Liu / Dan-Yuan Li / Ao Sun / Cheng-Ping Li / Shun-Qing Tan / Gao-Li Wang / Jie-Yi Shen / ...Authors: Zi-Xian Liu / Shouyue Zhang / Han-Zhou Zhu / Zhi-Hang Chen / Yun Yang / Long-Qi Li / Yuan Lei / Yun Liu / Dan-Yuan Li / Ao Sun / Cheng-Ping Li / Shun-Qing Tan / Gao-Li Wang / Jie-Yi Shen / Shuai Jin / Caixia Gao / Jun-Jie Gogo Liu /  Abstract: Ribozymes are catalytic RNAs with diverse functions including self-splicing and polymerization. This work aims to discover natural ribozymes that behave as hydrolytic and sequence-specific DNA ...Ribozymes are catalytic RNAs with diverse functions including self-splicing and polymerization. This work aims to discover natural ribozymes that behave as hydrolytic and sequence-specific DNA endonucleases, which could be repurposed as DNA manipulation tools. Focused on bacterial group II-C introns, we found that many systems without intron-encoded protein propagate multiple copies in their resident genomes. These introns, named HYdrolytic Endonucleolytic Ribozymes (HYERs), cleaved RNA, single-stranded DNA, bubbled double-stranded DNA (dsDNA), and plasmids in vitro. HYER1 generated dsDNA breaks in the mammalian genome. Cryo-electron microscopy analysis revealed a homodimer structure for HYER1, where each monomer contains a Mg-dependent hydrolysis pocket and captures DNA complementary to the target recognition site (TRS). Rational designs including TRS extension, recruiting sequence insertion, and heterodimerization yielded engineered HYERs showing improved specificity and flexibility for DNA manipulation. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_36779.map.gz emd_36779.map.gz | 450.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-36779-v30.xml emd-36779-v30.xml emd-36779.xml emd-36779.xml | 14.5 KB 14.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_36779.png emd_36779.png | 42.4 KB | ||
| Filedesc metadata |  emd-36779.cif.gz emd-36779.cif.gz | 5 KB | ||
| Others |  emd_36779_half_map_1.map.gz emd_36779_half_map_1.map.gz emd_36779_half_map_2.map.gz emd_36779_half_map_2.map.gz | 443 MB 443 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-36779 http://ftp.pdbj.org/pub/emdb/structures/EMD-36779 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36779 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36779 | HTTPS FTP |
-Related structure data
| Related structure data |  8k0sMC  8k0pC  8k0qC  8k0rC  8k15C M: atomic model generated by this map C: citing same article ( |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_36779.map.gz / Format: CCP4 / Size: 476.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_36779.map.gz / Format: CCP4 / Size: 476.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.0825 Å | ||||||||||||||||||||||||||||||||||||
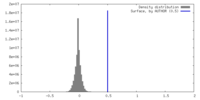
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_36779_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
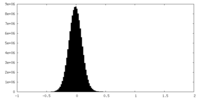
| Density Histograms |
-Half map: #1
| File | emd_36779_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : The Anoxybacillus pushchinoensis ORF-less Group IIC Intron HYER1 ...
| Entire | Name: The Anoxybacillus pushchinoensis ORF-less Group IIC Intron HYER1 with 10-nt TRS at symmetric apo state |
|---|---|
| Components |
|
-Supramolecule #1: The Anoxybacillus pushchinoensis ORF-less Group IIC Intron HYER1 ...
| Supramolecule | Name: The Anoxybacillus pushchinoensis ORF-less Group IIC Intron HYER1 with 10-nt TRS at symmetric apo state type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Anoxybacillus pushchinoensis (bacteria) Anoxybacillus pushchinoensis (bacteria) |
-Macromolecule #1: RNA (543-MER)
| Macromolecule | Name: RNA (543-MER) / type: rna / ID: 1 / Number of copies: 2 |
|---|---|
| Source (natural) | Organism:  Anoxybacillus pushchinoensis (bacteria) Anoxybacillus pushchinoensis (bacteria) |
| Molecular weight | Theoretical: 206.396656 KDa |
| Sequence | String: GUGCGCUCGG CAUGGGUGCA AUCUCUAGGG UGAAAGUCCC GAACUGCGAA GGCAGAAGUA GCAGUUAGCU UAACGCAAGG GUGUCCGUG GUGACGCGGA AUCUGAAGGA AGCGGGCGGC AAACUUCCGG UCUGAGGAAC ACGAACUUCA UAUAAGGCUA G GUAUCAUU ...String: GUGCGCUCGG CAUGGGUGCA AUCUCUAGGG UGAAAGUCCC GAACUGCGAA GGCAGAAGUA GCAGUUAGCU UAACGCAAGG GUGUCCGUG GUGACGCGGA AUCUGAAGGA AGCGGGCGGC AAACUUCCGG UCUGAGGAAC ACGAACUUCA UAUAAGGCUA G GUAUCAUU GGAUGAGUUU GCCGCUAAGA CAAAACAAAG UCCUUUCUGC CGAAGGUGAU ACAGAGUAAA UGAAGCAGAU AG AUGGAAG GAAAGAUUGU ACUCUUACCC GAGGAGGUCU GAUGGAUACG UGAAGUGCGC UUCAUAACCU ACUUAGUGAU AAG UAACUG AACCAUCAGA AGUCAGCAGA GGUCAUAGUA CGAAUCGGUC UAGAACGAUU CGGAAGGACU GAACAAUCAA GAGA AAAUA GCCCUUGGCA UUCAGUACGU CAUGAUGAAC ACAGAAAACA UGGUACCUCC CAAGAGAAAG GAAACGGUGA AUCCC GUGG GAAUCUUUUG GAGGGUGGAG UGACGACUGG CAUAAGAAGA UCAGCUAUUU ACGGAAGGAA GCUUGCGUCA UUAUCU UGA UUGAACCGCC GUAUACGGAA CCGUACGUAC GGUGGUGUGA GAGGACGGAG GUUAAUCACC UCCUCCUACU CGAU |
-Macromolecule #2: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 2 / Number of copies: 2 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: OTHER |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.41 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 166165 |
| Initial angle assignment | Type: OTHER |
| Final angle assignment | Type: OTHER |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)