-Search query
-Search result
Showing 1 - 50 of 3,744 items for (author: lei & d)

EMDB-37130: 
Cryo-EM structure of the human parainfluenza virus hPIV3 L-P polymerase in dimeric form
Method: single particle / : Xie J, Wang L, Zhai G, Wu D, Lin Z, Wang M, Yan X, Gao L, Huang X, Fearns R, Chen S

EMDB-37131: 
Cryo-EM structure of the human parainfluenza virus hPIV3 L-P polymerase in monomeric form
Method: single particle / : Xie J, Wang L, Zhai G, Wu D, Lin Z, Wang M, Yan X, Gao L, Huang X, Fearns R, Chen S

PDB-8kdb: 
Cryo-EM structure of the human parainfluenza virus hPIV3 L-P polymerase in dimeric form
Method: single particle / : Xie J, Wang L, Zhai G, Wu D, Lin Z, Wang M, Yan X, Gao L, Huang X, Fearns R, Chen S

PDB-8kdc: 
Cryo-EM structure of the human parainfluenza virus hPIV3 L-P polymerase in monomeric form
Method: single particle / : Xie J, Wang L, Zhai G, Wu D, Lin Z, Wang M, Yan X, Gao L, Huang X, Fearns R, Chen S

EMDB-18482: 
Herpes simplex virus 1 capsid (WT) vertices in perinuclear NEC-coated vesicles determined in situ
Method: subtomogram averaging / : Mironova Y, Prazak V, Vasishtan D

EMDB-18484: 
Herpes simplex virus 1 nuclear egress complex (WT) determined in situ from perinuclear vesicles
Method: subtomogram averaging / : Mironova Y, Prazak V, Vasishtan D

EMDB-40180: 
MsbA bound to cerastecin C
Method: single particle / : Chen Y, Klein D

EMDB-17766: 
CryoEM structure of Nal1 protein, allele SPIKE, from Oryza sativa japonica group
Method: single particle / : Huang LY, Rety S, Xi XG

EMDB-17768: 
CryoEM structure of Nal1 protein, allele IR64, from Oryza sativa indica cultivar
Method: single particle / : Huang LY, Rety S, Xi XG

PDB-8pn1: 
CryoEM structure of Nal1 protein, allele SPIKE, from Oryza sativa japonica group
Method: single particle / : Huang LY, Rety S, Xi XG

PDB-8pn2: 
CryoEM structure of Nal1 protein, allele IR64, from Oryza sativa indica cultivar
Method: single particle / : Huang LY, Rety S, Xi XG

EMDB-37240: 
SARS-CoV-2 Omicron spike in complex with 5817 Fab
Method: single particle / : Cao L, Wang X

EMDB-37241: 
The interface structure of Omicron RBD binding to 5817 Fab
Method: single particle / : Cao L, Wang X

PDB-8khc: 
SARS-CoV-2 Omicron spike in complex with 5817 Fab
Method: single particle / : Cao L, Wang X

PDB-8khd: 
The interface structure of Omicron RBD binding to 5817 Fab
Method: single particle / : Cao L, Wang X

EMDB-17974: 
Pseudorabies virus cytosolic C-capsid (US3 KO) vertices determined in situ
Method: subtomogram averaging / : Prazak V, Grange M, Vasishtan D

EMDB-17975: 
Pseudorabies virus primary enveloped (perinuclear) C-capsid (US3 KO) vertices determined in situ
Method: subtomogram averaging / : Prazak V, Grange M, Vasishtan D

EMDB-17976: 
Pseudorabies nuclear C-capsids (US3 KO) vertices determined in situ
Method: subtomogram averaging / : Prazak V, Grange M, Vasishtan D

EMDB-18473: 
Subtomogram average of pseudorabies virus nuclear egress complex helical form (UL31/34) determined in situ
Method: subtomogram averaging / : Prazak V, Grange M, Vasishtan D

EMDB-18474: 
Subtomogram average of pseudorabies virus nuclear egress complex (UL31/34) determined in situ
Method: subtomogram averaging / : Prazak V, Grange M, Vasishtan D

EMDB-18479: 
Pseudorabies virus cytosolic C-capsid (WT) vertices determined in situ
Method: subtomogram averaging / : Mironova Y, Prazak V, Vasishtan D

EMDB-18480: 
Pseudorabies virus nuclear C-capsid (WT) vertices determined in situ
Method: subtomogram averaging / : Mironova Y, Prazak V, Vasishtan D

EMDB-18481: 
Herpes simplex virus 1 cytosolic C-capsid (WT) vertices determined in situ
Method: subtomogram averaging / : Mironova Y, Prazak V, Vasishtan D

EMDB-18483: 
Herpes simplex virus 1 nuclear C-capsid (WT) vertices determined in situ
Method: subtomogram averaging / : Mironova Y, Prazak V, Vasishtan D

EMDB-19856: 
Focused map 1- K48-linked ubiquitin chain formation with a cullin-RING E3 ligase & Cdc34: NEDD8-CUL2-RBX1-ELOB/C-FEM1C with trapped UBE2R2~donor UB~acceptor UB-SIL1 peptide
Method: single particle / : Liwocha J, Prabu JR, Kleiger G, Schulman BA

EMDB-19857: 
Focused map 2 - K48-linked ubiquitin chain formation with a cullin-RING E3 ligase & Cdc34: NEDD8-CUL2-RBX1-ELOB/C-FEM1C with trapped UBE2R2~donor UB~acceptor UB-SIL1 peptide
Method: single particle / : Liwocha J, Prabu JR, Kleiger G, Schulman BA

EMDB-19858: 
Focused map 3 - K48-linked ubiquitin chain formation with a cullin-RING E3 ligase & Cdc34: NEDD8-CUL2-RBX1-ELOB/C-FEM1C with trapped UBE2R2~donor UB~acceptor UB-SIL1 peptide
Method: single particle / : Liwocha J, Prabu JR, Kleiger G, Schulman BA

EMDB-19859: 
Focused map 4 - K48-linked ubiquitin chain formation with a cullin-RING E3 ligase & Cdc34: NEDD8-CUL2-RBX1-ELOB/C-FEM1C with trapped UBE2R2~donor UB~acceptor UB-SIL1 peptide
Method: single particle / : Liwocha J, Prabu JR, Kleiger G, Schulman BA

EMDB-19860: 
Focused map 5 - K48-linked ubiquitin chain formation with a cullin-RING E3 ligase & Cdc34: NEDD8-CUL2-RBX1-ELOB/C-FEM1C with trapped UBE2R2~donor UB~acceptor UB-SIL1 peptide
Method: single particle / : Liwocha J, Prabu JR, Kleiger G, Schulman BA
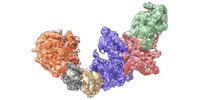
EMDB-36808: 
Cryo-EM structure of KEOPS complex from Arabidopsis thaliana
Method: single particle / : Zheng XX, Zhu L, Duan L, Zhang WH

PDB-8k20: 
Cryo-EM structure of KEOPS complex from Arabidopsis thaliana
Method: single particle / : Zheng XX, Zhu L, Duan L, Zhang WH

EMDB-18110: 
The fibrillar and amorphous states of polyQ Q97
Method: electron tomography / : Zhao DY

EMDB-18114: 
phagophore in fibrillar polyQ
Method: electron tomography / : Zhao DY
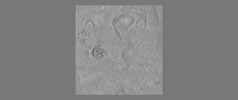
EMDB-18115: 
phagophore and lysosomes with amorphous polyQ
Method: electron tomography / : Zhao DY
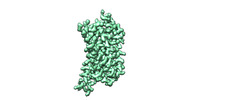
EMDB-43732: 
momSalB bound Kappa Opioid Receptor in complex Gi1
Method: single particle / : Fay JF, Che T

EMDB-43733: 
GR89,696 bound Kappa Opioid Receptor in complex with Gz
Method: single particle / : Fay JF, Che T

EMDB-43734: 
GR89,696 bound Kappa Opioid Receptor in complex with gustducin
Method: single particle / : Fay JF, Che T

EMDB-43715: 
HTT in complex with HAP40 in the apo state.
Method: single particle / : Poweleit N, Boudet J, Doherty E

PDB-8w15: 
HTT in complex with HAP40 in the apo state.
Method: single particle / : Poweleit N, Boudet J, Doherty E

EMDB-41888: 
Structure of Apo CXCR4/Gi complex
Method: single particle / : Saotome K, McGoldrick LL, Franklin MC

EMDB-41889: 
Structure of CXCL12-bound CXCR4/Gi complex
Method: single particle / : Saotome K, McGoldrick LL, Franklin MC

EMDB-41890: 
Structure of AMD3100-bound CXCR4/Gi complex
Method: single particle / : Saotome K, McGoldrick LL, Franklin MC

EMDB-41891: 
Structure of REGN7663 Fab-bound CXCR4/Gi complex
Method: single particle / : Saotome K, McGoldrick LL, Franklin MC

EMDB-41892: 
Structure of REGN7663-Fab bound CXCR4
Method: single particle / : Saotome K, McGoldrick LL, Franklin MC

EMDB-41893: 
Structure of trimeric CXCR4 in complex with REGN7663 Fab
Method: single particle / : Saotome K, McGoldrick LL, Franklin MC

EMDB-41894: 
Structure of tetrameric CXCR4 in complex with REGN7663 Fab
Method: single particle / : Saotome K, McGoldrick LL, Franklin MC

EMDB-42681: 
The structure of the native cardiac thin filament troponin core in Ca2+-free state from the upper strand
Method: single particle / : Galkin VE, Risi CM

EMDB-42682: 
The structure of the native cardiac thin filament troponin core in Ca2+-free tilted state from the upper strand
Method: single particle / : Galkin VE, Risi CM

EMDB-42683: 
The structure of the native cardiac thin filament troponin core in Ca2+-free rotated state from the upper strand
Method: single particle / : Galkin VE, Risi CM
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search




 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
