[English] 日本語
 Yorodumi
Yorodumi- EMDB-18484: Herpes simplex virus 1 nuclear egress complex (WT) determined in ... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Herpes simplex virus 1 nuclear egress complex (WT) determined in situ from perinuclear vesicles | ||||||||||||||||||
 Map data Map data | Herpes simplex virus-1 nuclear egress complex determined in situ | ||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||
 Keywords Keywords | Capsid / capsid vertex specific component / in situ / virus | ||||||||||||||||||
| Biological species |   Human alphaherpesvirus 1 (Herpes simplex virus type 1) Human alphaherpesvirus 1 (Herpes simplex virus type 1) | ||||||||||||||||||
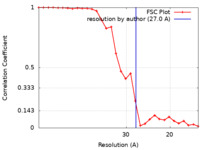
| Method | subtomogram averaging / cryo EM / Resolution: 27.0 Å | ||||||||||||||||||
 Authors Authors | Mironova Y / Prazak V / Vasishtan D | ||||||||||||||||||
| Funding support |  United Kingdom, United Kingdom,  Germany, 5 items Germany, 5 items
| ||||||||||||||||||
 Citation Citation |  Journal: Nat Microbiol / Year: 2024 Journal: Nat Microbiol / Year: 2024Title: Molecular plasticity of herpesvirus nuclear egress analysed in situ. Authors: Vojtěch Pražák / Yuliia Mironova / Daven Vasishtan / Christoph Hagen / Ulrike Laugks / Yannick Jensen / Saskia Sanders / John M Heumann / Jens B Bosse / Barbara G Klupp / Thomas C ...Authors: Vojtěch Pražák / Yuliia Mironova / Daven Vasishtan / Christoph Hagen / Ulrike Laugks / Yannick Jensen / Saskia Sanders / John M Heumann / Jens B Bosse / Barbara G Klupp / Thomas C Mettenleiter / Michael Grange / Kay Grünewald /    Abstract: The viral nuclear egress complex (NEC) allows herpesvirus capsids to escape from the nucleus without compromising the nuclear envelope integrity. The NEC lattice assembles on the inner nuclear ...The viral nuclear egress complex (NEC) allows herpesvirus capsids to escape from the nucleus without compromising the nuclear envelope integrity. The NEC lattice assembles on the inner nuclear membrane and mediates the budding of nascent nucleocapsids into the perinuclear space and their subsequent release into the cytosol. Its essential role makes it a potent antiviral target, necessitating structural information in the context of a cellular infection. Here we determined structures of NEC-capsid interfaces in situ using electron cryo-tomography, showing a substantial structural heterogeneity. In addition, while the capsid is associated with budding initiation, it is not required for curvature formation. By determining the NEC structure in several conformations, we show that curvature arises from an asymmetric assembly of disordered and hexagonally ordered lattice domains independent of pUL25 or other viral capsid vertex components. Our results advance our understanding of the mechanism of nuclear egress in the context of a living cell. | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_18484.map.gz emd_18484.map.gz | 677.5 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-18484-v30.xml emd-18484-v30.xml emd-18484.xml emd-18484.xml | 17.6 KB 17.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_18484_fsc.xml emd_18484_fsc.xml | 2.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_18484.png emd_18484.png | 43.9 KB | ||
| Filedesc metadata |  emd-18484.cif.gz emd-18484.cif.gz | 4.8 KB | ||
| Others |  emd_18484_additional_1.map.gz emd_18484_additional_1.map.gz emd_18484_half_map_1.map.gz emd_18484_half_map_1.map.gz emd_18484_half_map_2.map.gz emd_18484_half_map_2.map.gz | 4.2 MB 697.8 KB 697.9 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-18484 http://ftp.pdbj.org/pub/emdb/structures/EMD-18484 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18484 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18484 | HTTPS FTP |
-Related structure data
| Related structure data | C: citing same article ( |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_18484.map.gz / Format: CCP4 / Size: 768.6 KB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_18484.map.gz / Format: CCP4 / Size: 768.6 KB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Herpes simplex virus-1 nuclear egress complex determined in situ | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 8.484 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: Unfiltered map at bin4
| File | emd_18484_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unfiltered map at bin4 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map 1
| File | emd_18484_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map 2
| File | emd_18484_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Human alphaherpesvirus 1
| Entire | Name:   Human alphaherpesvirus 1 (Herpes simplex virus type 1) Human alphaherpesvirus 1 (Herpes simplex virus type 1) |
|---|---|
| Components |
|
-Supramolecule #1: Human alphaherpesvirus 1
| Supramolecule | Name: Human alphaherpesvirus 1 / type: virus / ID: 1 / Parent: 0 / NCBI-ID: 10298 / Sci species name: Human alphaherpesvirus 1 / Sci species strain: 17 / Virus type: VIRION / Virus isolate: STRAIN / Virus enveloped: No / Virus empty: No |
|---|
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | cell |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Grid | Model: Quantifoil R2/2 / Material: GOLD / Support film - topology: HOLEY |
| Vitrification | Cryogen name: ETHANE-PROPANE / Chamber humidity: 70 % / Chamber temperature: 310.15 K Details: Vitrified samples were milled using dual beam cryo-FIB-SEM (Aquilos). |
| Details | Vero cells were infected with HSV1 (strain 17) at MOI 30 and incubated for 15 h |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average exposure time: 0.4 sec. / Average electron dose: 2.5 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 6.0 µm / Nominal defocus min: 5.0 µm / Nominal magnification: 26000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)