-Search query
-Search result
Showing 1 - 50 of 63 items for (author: hsia & rc)

EMDB-27031: 
Accurate computational design of genetically encoded 3D protein crystals
Method: single particle / : Li Z, Borst AJ, Baker D

EMDB-40926: 
CryoEM Structure of Computationally Designed Nanocage O32-ZL4
Method: single particle / : Weidle C, Borst A

PDB-8cwy: 
Accurate computational design of genetically encoded 3D protein crystals
Method: single particle / : Li Z, Borst AJ, Baker D

PDB-8szz: 
CryoEM Structure of Computationally Designed Nanocage O32-ZL4
Method: single particle / : Weidle C, Borst A

EMDB-29725: 
Vaccine-elicited human antibody 2C06 in complex with HIV-1 envelope trimer BG505 DS-SOSIP
Method: single particle / : Wang S, Morano NC, Shapiro L, Kwong PD

EMDB-29731: 
Vaccine-elicited human antibody 2C09 in complex with HIV-1 envelope trimer BG505 DS-SOSIP
Method: single particle / : Wang S, Kwong PD

PDB-8g4m: 
Vaccine-elicited human antibody 2C06 in complex with HIV-1 envelope trimer BG505 DS-SOSIP
Method: single particle / : Wang S, Morano NC, Shapiro L, Kwong PD

PDB-8g4t: 
Vaccine-elicited human antibody 2C09 in complex with HIV-1 envelope trimer BG505 DS-SOSIP
Method: single particle / : Wang S, Kwong PD
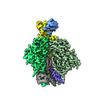
EMDB-29248: 
Cryo-EM Structure of PG9RSH DU011 Fab in complex with BG505 DS-SOSIP.664
Method: single particle / : Gorman J, Kwong PD
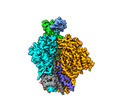
EMDB-29264: 
Cryo-EM Structure of PG9RSH DU025 Fab in complex with BG505 DS-SOSIP.664
Method: single particle / : Gorman J, Kwong PD
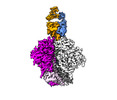
EMDB-29288: 
Cryo-EM Structure of PGT145 DU303 Fab in complex with BG505 DS-SOSIP.664
Method: single particle / : Gorman J, Kwong PD

PDB-8fk5: 
Cryo-EM Structure of PG9RSH DU011 Fab in complex with BG505 DS-SOSIP.664
Method: single particle / : Gorman J, Kwong PD

PDB-8fl1: 
Cryo-EM Structure of PG9RSH DU025 Fab in complex with BG505 DS-SOSIP.664
Method: single particle / : Gorman J, Kwong PD

PDB-8flw: 
Cryo-EM Structure of PGT145 DU303 Fab in complex with BG505 DS-SOSIP.664
Method: single particle / : Gorman J, Kwong PD

EMDB-27148: 
Zebrafish MFSD2A isoform B in inward open ligand bound conformation
Method: single particle / : Nguyen C, Lei HT, Lai LTF, Gallentino MJ, Mu X, Matthies D, Gonen T

EMDB-27149: 
Zebrafish MFSD2A isoform B in inward open ligand-free conformation
Method: single particle / : Nguyen C, Lei HT, Lai LTF, Gallentino MJ, Mu X, Matthies D, Gonen T

EMDB-27150: 
Zebrafish MFSD2A isoform B in inward open ligand 1A conformation
Method: single particle / : Nguyen C, Lei HT, Lai LTF, Gallentino MJ, Mu X, Matthies D, Gonen T

EMDB-27151: 
Zebrafish MFSD2A isoform B in inward open ligand 1B conformation
Method: single particle / : Nguyen C, Lei HT, Lai LTF, Gallentino MJ, Mu X, Matthies D, Gonen T

EMDB-27152: 
Zebrafish MFSD2A isoform B in inward open ligand 2B conformation
Method: single particle / : Nguyen C, Lei HT, Lai LTF, Gallentino MJ, Mu X, Matthies D, Gonen T

EMDB-27153: 
Zebrafish MFSD2A isoform B in inward open ligand 3C conformation
Method: single particle / : Nguyen C, Lei HT, Lai LTF, Gallentino MJ, Mu X, Matthies D, Gonen T

PDB-8d2s: 
Zebrafish MFSD2A isoform B in inward open ligand bound conformation
Method: single particle / : Nguyen C, Lei HT, Lai LTF, Gallentino MJ, Mu X, Matthies D, Gonen T

PDB-8d2t: 
Zebrafish MFSD2A isoform B in inward open ligand-free conformation
Method: single particle / : Nguyen C, Lei HT, Lai LTF, Gallentino MJ, Mu X, Matthies D, Gonen T

PDB-8d2u: 
Zebrafish MFSD2A isoform B in inward open ligand 1A conformation
Method: single particle / : Nguyen C, Lei HT, Lai LTF, Gallentino MJ, Mu X, Matthies D, Gonen T

PDB-8d2v: 
Zebrafish MFSD2A isoform B in inward open ligand 1B conformation
Method: single particle / : Nguyen C, Lei HT, Lai LTF, Gallentino MJ, Mu X, Matthies D, Gonen T

PDB-8d2w: 
Zebrafish MFSD2A isoform B in inward open ligand 2B conformation
Method: single particle / : Nguyen C, Lei HT, Lai LTF, Gallentino MJ, Mu X, Matthies D, Gonen T

PDB-8d2x: 
Zebrafish MFSD2A isoform B in inward open ligand 3C conformation
Method: single particle / : Nguyen C, Lei HT, Lai LTF, Gallentino MJ, Mu X, Matthies D, Gonen T

EMDB-29396: 
Antibody vFP53.02 in complex with HIV-1 envelope trimer BG505 DS-SOSIP
Method: single particle / : Wang S, Kwong PD
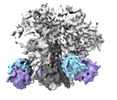
EMDB-29836: 
vFP52.02 Fab in complex with BG505 DS-SOSIP Env trimer
Method: single particle / : Gorman J, Kwong PD

EMDB-29880: 
Cryo-EM structure of vFP49.02 Fab in complex with HIV-1 Env BG505 DS-SOSIP.664 (conformation 1)
Method: single particle / : Changela A, Gorman J, Kwong PD

EMDB-29881: 
Cryo-EM structure of vFP49.02 Fab in complex with HIV-1 Env BG505 DS-SOSIP.664 (conformation 2)
Method: single particle / : Changela A, Gorman J, Kwong PD

EMDB-29882: 
Cryo-EM structure of vFP49.02 Fab in complex with HIV-1 Env BG505 DS-SOSIP.664 (conformation 3)
Method: single particle / : Changela A, Gorman J, Kwong PD

EMDB-29905: 
vFP48.02 Fab in complex with BG505 DS-SOSIP Env trimer
Method: single particle / : Gorman J, Kwong PD

PDB-8fr6: 
Antibody vFP53.02 in complex with HIV-1 envelope trimer BG505 DS-SOSIP
Method: single particle / : Wang S, Kwong PD

PDB-8g85: 
vFP52.02 Fab in complex with BG505 DS-SOSIP Env trimer
Method: single particle / : Gorman J, Kwong PD

PDB-8g9w: 
Cryo-EM structure of vFP49.02 Fab in complex with HIV-1 Env BG505 DS-SOSIP.664 (conformation 1)
Method: single particle / : Changela A, Gorman J, Kwong PD

PDB-8g9x: 
Cryo-EM structure of vFP49.02 Fab in complex with HIV-1 Env BG505 DS-SOSIP.664 (conformation 2)
Method: single particle / : Changela A, Gorman J, Kwong PD

PDB-8g9y: 
Cryo-EM structure of vFP49.02 Fab in complex with HIV-1 Env BG505 DS-SOSIP.664 (conformation 3)
Method: single particle / : Changela A, Gorman J, Kwong PD

PDB-8gas: 
vFP48.02 Fab in complex with BG505 DS-SOSIP Env trimer
Method: single particle / : Gorman J, Kwong PD

EMDB-28862: 
KWOCA 4 nanoparticle
Method: single particle / : Antanasijevic A, Ward AB

EMDB-28929: 
Improving the secretion of designed protein assemblies through negative design of cryptic transmembrane domains - KWOCA51
Method: single particle / : Borst AJ, King NP

EMDB-23098: 
Cryo-EM structure of the VRC316 clinical trial, vaccine-elicited, human antibody 316-310-1B11 in complex with an H2 CAN05 HA trimer
Method: single particle / : Gorman J, Kwong PD

EMDB-23816: 
Cryo-EM structure of the VRC310 clinical trial, vaccine-elicited, human antibody 310-030-1D06 Fab in complex with an H1 NC99 HA trimer
Method: single particle / : Gorman J, Kwong PD

PDB-7l0l: 
Cryo-EM structure of the VRC316 clinical trial, vaccine-elicited, human antibody 316-310-1B11 in complex with an H2 CAN05 HA trimer
Method: single particle / : Gorman J, Kwong PD

PDB-7mfg: 
Cryo-EM structure of the VRC310 clinical trial, vaccine-elicited, human antibody 310-030-1D06 Fab in complex with an H1 NC99 HA trimer
Method: single particle / : Gorman J, Kwong PD

EMDB-21961: 
Cryo-EM structure of the VRC315 clinical trial, vaccine-elicited, human antibody 1D12 in complex with an H7 SH13 HA trimer
Method: single particle / : Gorman J, Kwong PD

PDB-6wxl: 
Cryo-EM structure of the VRC315 clinical trial, vaccine-elicited, human antibody 1D12 in complex with an H7 SH13 HA trimer
Method: single particle / : Gorman J, Kwong PD

EMDB-22331: 
SPN3US phage empty capsid
Method: single particle / : Heymann JB, Wang B, Newcomb WW, Wu W, Winkler DC, Cheng N, Reilly ER, Hsia RC, Thomas JA, Steven AC

EMDB-22332: 
Phage SPN3US mottled capsid (glutaraldehyde-fixed)
Method: single particle / : Heymann JB, Wang B, Newcomb WW, Wu W, Winkler DC, Cheng N, Reilly ER, Hsia RC, Thomas JA, Steven AC

EMDB-22333: 
SPN3US phage mottled capsid
Method: single particle / : Heymann JB, Wang B, Newcomb WW, Wu W, Winkler DC, Cheng N, Reilly ER, Hsia RC, Thomas JA, Steven AC

EMDB-8977: 
Cryo-EM structure at 3.2 A resolution of HIV-1 fusion peptide-directed antibody, A12V163-b.01, elicited by vaccination of Rhesus macaques, in complex with stabilized HIV-1 Env BG505 DS-SOSIP, which was also bound to antibodies VRC03 and PGT122
Method: single particle / : Acharya P, Eng ET, Kwong PD
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
