[English] 日本語
 Yorodumi
Yorodumi- EMDB-23098: Cryo-EM structure of the VRC316 clinical trial, vaccine-elicited,... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-23098 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
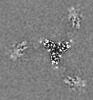
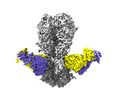
| Title | Cryo-EM structure of the VRC316 clinical trial, vaccine-elicited, human antibody 316-310-1B11 in complex with an H2 CAN05 HA trimer | |||||||||
 Map data Map data | Sharpened map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | VRC / IMMUNE SYSTEM / VRC316 / H2 / Fab / Flu / IMMUNE SYSTEM-Viral protein complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationviral budding from plasma membrane / clathrin-dependent endocytosis of virus by host cell / host cell surface receptor binding / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / virion attachment to host cell / host cell plasma membrane / virion membrane / membrane Similarity search - Function | |||||||||
| Biological species |   Influenza A virus / Influenza A virus /  Homo sapiens (human) / Homo sapiens (human) /  Influenza A virus (A/Canada/720/2005(H2N2)) Influenza A virus (A/Canada/720/2005(H2N2)) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.85 Å | |||||||||
 Authors Authors | Gorman J / Kwong PD | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Med / Year: 2022 Journal: Nat Med / Year: 2022Title: A single residue in influenza virus H2 hemagglutinin enhances the breadth of the B cell response elicited by H2 vaccination. Authors: Sarah F Andrews / Julie E Raab / Jason Gorman / Rebecca A Gillespie / Crystal S F Cheung / Reda Rawi / Lauren Y Cominsky / Jeffrey C Boyington / Adrian Creanga / Chen-Hsiang Shen / Darcy R ...Authors: Sarah F Andrews / Julie E Raab / Jason Gorman / Rebecca A Gillespie / Crystal S F Cheung / Reda Rawi / Lauren Y Cominsky / Jeffrey C Boyington / Adrian Creanga / Chen-Hsiang Shen / Darcy R Harris / Adam S Olia / Alexandra F Nazzari / Tongqing Zhou / Katherine V Houser / Grace L Chen / John R Mascola / Barney S Graham / Masaru Kanekiyo / Julie E Ledgerwood / Peter D Kwong / Adrian B McDermott /  Abstract: Conserved epitopes on the influenza hemagglutinin (HA) stem are an attractive target for universal vaccine strategies as they elicit broadly neutralizing antibodies. Such antibody responses to stem- ...Conserved epitopes on the influenza hemagglutinin (HA) stem are an attractive target for universal vaccine strategies as they elicit broadly neutralizing antibodies. Such antibody responses to stem-specific epitopes have been extensively characterized for HA subtypes H1 and H5 in humans. H2N2 influenza virus circulated 50 years ago and represents a pandemic threat due to the lack of widespread immunity, but, unlike H1 and H5, the H2 HA stem contains Phe45 predicted to sterically clash with HA stem-binding antibodies characterized to date. To understand the effect of Phe45, we compared the HA stem-specific B cell response in post hoc analyses of two phase 1 clinical trials, one testing vaccination with an H2 ferritin nanoparticle immunogen ( NCT03186781 ) and one with an inactivated H5N1 vaccine ( NCT01086657 ). In H2-naive individuals, the magnitude of the B cell response was equivalent, but H2-elicited HA stem-binding B cells displayed greater cross-reactivity than those elicited by H5. However, in individuals with childhood H2 exposure, H5-elicited HA stem-binding B cells also displayed high cross-reactivity, suggesting recall of memory B cells formed 50 years ago. Overall, we propose that a one-residue difference on an HA immunogen can alter establishment and expansion of broadly neutralizing memory B cells. These data have implications for stem-based universal influenza vaccination strategies. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_23098.map.gz emd_23098.map.gz | 116.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-23098-v30.xml emd-23098-v30.xml emd-23098.xml emd-23098.xml | 25.5 KB 25.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_23098.png emd_23098.png | 119.3 KB | ||
| Masks |  emd_23098_msk_1.map emd_23098_msk_1.map | 125 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-23098.cif.gz emd-23098.cif.gz | 7.1 KB | ||
| Others |  emd_23098_additional_1.map.gz emd_23098_additional_1.map.gz emd_23098_additional_2.map.gz emd_23098_additional_2.map.gz emd_23098_half_map_1.map.gz emd_23098_half_map_1.map.gz emd_23098_half_map_2.map.gz emd_23098_half_map_2.map.gz | 31.3 MB 16.8 MB 116 MB 116 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-23098 http://ftp.pdbj.org/pub/emdb/structures/EMD-23098 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23098 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23098 | HTTPS FTP |
-Related structure data
| Related structure data |  7l0lMC  7mfgC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_23098.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_23098.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.083 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
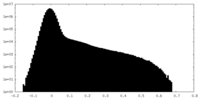
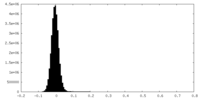
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_23098_msk_1.map emd_23098_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: unsharpened map
| File | emd_23098_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | unsharpened map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: density modified map with resolve
| File | emd_23098_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | density modified map with resolve | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map A
| File | emd_23098_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map B
| File | emd_23098_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : 316-310-1B11 Fab in complex with an H2 CAN05 HA trimer
| Entire | Name: 316-310-1B11 Fab in complex with an H2 CAN05 HA trimer |
|---|---|
| Components |
|
-Supramolecule #1: 316-310-1B11 Fab in complex with an H2 CAN05 HA trimer
| Supramolecule | Name: 316-310-1B11 Fab in complex with an H2 CAN05 HA trimer type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 |
|---|---|
| Source (natural) | Organism:   Influenza A virus Influenza A virus |
-Macromolecule #1: 316-310-1B11 Light Chain
| Macromolecule | Name: 316-310-1B11 Light Chain / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 23.460025 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: EIVLTQSPGT LSLSPGDRAT LSCRASQSVP SSYLAWYRHK PGQAPRLLIY GASSRATGIP DRFSGSGSGT DFSLTISRVE PEDFAVYYC QQYGSSPYTF GRGTKLDIKR TVAAPSVFIF PPSDEQLKSG TASVVCLLNN FYPREAKVQW KVDNALQSGN S QESVTEQD ...String: EIVLTQSPGT LSLSPGDRAT LSCRASQSVP SSYLAWYRHK PGQAPRLLIY GASSRATGIP DRFSGSGSGT DFSLTISRVE PEDFAVYYC QQYGSSPYTF GRGTKLDIKR TVAAPSVFIF PPSDEQLKSG TASVVCLLNN FYPREAKVQW KVDNALQSGN S QESVTEQD SKDSTYSLSS TLTLSKADYE KHKVYACEVT HQGLSSPVTK SFNRGEC |
-Macromolecule #2: Hemagglutinin HA1 chain
| Macromolecule | Name: Hemagglutinin HA1 chain / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Influenza A virus (A/Canada/720/2005(H2N2)) / Strain: A/Canada/720/2005(H2N2) Influenza A virus (A/Canada/720/2005(H2N2)) / Strain: A/Canada/720/2005(H2N2) |
| Molecular weight | Theoretical: 38.802152 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MAIIYLILLF TAVRGDQICI GYHANNSTEK VDTICERNVT VTHAKDILEK THNGKLCKLN GIPPLELGDC SIAGWLLGNP ECDRLLSVP EWSYIMEKEN PRDGLCYPGS FNDYEELKHL LSSVKHFEKV KILPKDRWTQ HTTTGGSRAC AVSGNPSFFR N MVWLTKKG ...String: MAIIYLILLF TAVRGDQICI GYHANNSTEK VDTICERNVT VTHAKDILEK THNGKLCKLN GIPPLELGDC SIAGWLLGNP ECDRLLSVP EWSYIMEKEN PRDGLCYPGS FNDYEELKHL LSSVKHFEKV KILPKDRWTQ HTTTGGSRAC AVSGNPSFFR N MVWLTKKG SNYPVAQGSY NNTSGEQMLI IWGVHHPNDE TEQRTLYQNV GTYVSVGTST LNKRSTPEIA TRPKVNGQGG RM EFSWTLL DMWDTINFES TGNLIAPEYG FKISKRGSSG IMKTEGTLEN CETKCQTPLG AINTTLPFHN VHPLTIGECP KYV KSEKLV LATGLRNVPQ IESRRRRRR UniProtKB: Hemagglutinin |
-Macromolecule #3: Hemagglutinin HA2 chain
| Macromolecule | Name: Hemagglutinin HA2 chain / type: protein_or_peptide / ID: 3 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Influenza A virus (A/Canada/720/2005(H2N2)) / Strain: A/Canada/720/2005(H2N2) Influenza A virus (A/Canada/720/2005(H2N2)) / Strain: A/Canada/720/2005(H2N2) |
| Molecular weight | Theoretical: 25.489357 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: GLFGAIAGFI EGGWQGMVDG WYGYHHSNDQ GSGYAADKES TQKAFDCITN KVNSVIEKMN TQFEAVGKEF SNLERRLENL NKKMEDGFL DVWTYNAELL VLMENERTLD FHDSNVKNLY DKVRMQLRDN VKELGNGCFE FYHKCDDECM NSVKNGTYDY P KYEEESKL ...String: GLFGAIAGFI EGGWQGMVDG WYGYHHSNDQ GSGYAADKES TQKAFDCITN KVNSVIEKMN TQFEAVGKEF SNLERRLENL NKKMEDGFL DVWTYNAELL VLMENERTLD FHDSNVKNLY DKVRMQLRDN VKELGNGCFE FYHKCDDECM NSVKNGTYDY P KYEEESKL NRNEIKGVKG RLVPRGSPGS GYIPEAPRDG QAYVRKDGEW VLLSTFLGHH HHHH UniProtKB: Hemagglutinin |
-Macromolecule #4: 316-310-1B11 Heavy Chain
| Macromolecule | Name: 316-310-1B11 Heavy Chain / type: protein_or_peptide / ID: 4 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 25.065092 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QVQLVQSGAE VKKPGSSVKV SCKTSGGIFR SNAISWVRQA PGQGLEWMGG VVAIFGTTNY AQNFQGRVTI TADESSSTVY MELSRLRSE DTAVYYCARH SGYHITNTFF DYWGQGTLVT VSSASTKGPS VFPLAPSSKS TSGGTAALGC LVKDYFPEPV T VSWNSGAL ...String: QVQLVQSGAE VKKPGSSVKV SCKTSGGIFR SNAISWVRQA PGQGLEWMGG VVAIFGTTNY AQNFQGRVTI TADESSSTVY MELSRLRSE DTAVYYCARH SGYHITNTFF DYWGQGTLVT VSSASTKGPS VFPLAPSSKS TSGGTAALGC LVKDYFPEPV T VSWNSGAL TSGVHTFPAV LQSSGLYSLS SVVTVPSSSL GTQTYICNVN HKPSNTKVDK KVEPKSCDKG LEVLFQGP |
-Macromolecule #5: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 5 / Number of copies: 15 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2 mg/mL |
|---|---|
| Buffer | pH: 7.4 / Component - Formula: PBS |
| Grid | Model: C-flat-1.2/1.3 / Material: COPPER / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: PLASMA CLEANING |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Chamber temperature: 293 K / Instrument: FEI VITROBOT MARK IV |
| Details | Cryo-EM structure of the VRC316 clinical trial, vaccine-elicited, human antibody 316-310-1B11 in complex with an H2 CAN05 HA trimer |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Detector mode: COUNTING / Number grids imaged: 1 / Number real images: 2439 / Average electron dose: 51.16 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)