[English] 日本語
 Yorodumi
Yorodumi- EMDB-28929: Improving the secretion of designed protein assemblies through ne... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Improving the secretion of designed protein assemblies through negative design of cryptic transmembrane domains - KWOCA51 | |||||||||
 Map data Map data | Unsharpened Map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | tetradron / KWOCA / secretable / DE NOVO PROTEIN / design / degreaser / cage / vaccine | |||||||||
| Biological species | synthetic construct (others) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 5.87 Å | |||||||||
 Authors Authors | Borst AJ / King NP | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2023 Journal: Proc Natl Acad Sci U S A / Year: 2023Title: Improving the secretion of designed protein assemblies through negative design of cryptic transmembrane domains. Authors: Jing Yang John Wang / Alena Khmelinskaia / William Sheffler / Marcos C Miranda / Aleksandar Antanasijevic / Andrew J Borst / Susana V Torres / Chelsea Shu / Yang Hsia / Una Nattermann / ...Authors: Jing Yang John Wang / Alena Khmelinskaia / William Sheffler / Marcos C Miranda / Aleksandar Antanasijevic / Andrew J Borst / Susana V Torres / Chelsea Shu / Yang Hsia / Una Nattermann / Daniel Ellis / Carl Walkey / Maggie Ahlrichs / Sidney Chan / Alex Kang / Hannah Nguyen / Claire Sydeman / Banumathi Sankaran / Mengyu Wu / Asim K Bera / Lauren Carter / Brooke Fiala / Michael Murphy / David Baker / Andrew B Ward / Neil P King /   Abstract: Computationally designed protein nanoparticles have recently emerged as a promising platform for the development of new vaccines and biologics. For many applications, secretion of designed ...Computationally designed protein nanoparticles have recently emerged as a promising platform for the development of new vaccines and biologics. For many applications, secretion of designed nanoparticles from eukaryotic cells would be advantageous, but in practice, they often secrete poorly. Here we show that designed hydrophobic interfaces that drive nanoparticle assembly are often predicted to form cryptic transmembrane domains, suggesting that interaction with the membrane insertion machinery could limit efficient secretion. We develop a general computational protocol, the Degreaser, to design away cryptic transmembrane domains without sacrificing protein stability. The retroactive application of the Degreaser to previously designed nanoparticle components and nanoparticles considerably improves secretion, and modular integration of the Degreaser into design pipelines results in new nanoparticles that secrete as robustly as naturally occurring protein assemblies. Both the Degreaser protocol and the nanoparticles we describe may be broadly useful in biotechnological applications. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_28929.map.gz emd_28929.map.gz | 83.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-28929-v30.xml emd-28929-v30.xml emd-28929.xml emd-28929.xml | 16.3 KB 16.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_28929.png emd_28929.png | 83 KB | ||
| Filedesc metadata |  emd-28929.cif.gz emd-28929.cif.gz | 4.8 KB | ||
| Others |  emd_28929_additional_1.map.gz emd_28929_additional_1.map.gz emd_28929_half_map_1.map.gz emd_28929_half_map_1.map.gz emd_28929_half_map_2.map.gz emd_28929_half_map_2.map.gz | 87.6 MB 164.9 MB 164.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-28929 http://ftp.pdbj.org/pub/emdb/structures/EMD-28929 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28929 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28929 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_28929.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_28929.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened Map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.84 Å | ||||||||||||||||||||||||||||||||||||
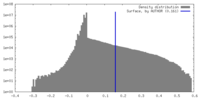
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: Sharpened Map
| File | emd_28929_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened Map | ||||||||||||
| Projections & Slices |
| ||||||||||||
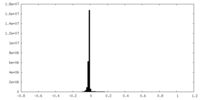
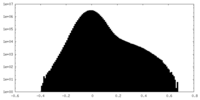
| Density Histograms |
-Half map: Half Map A
| File | emd_28929_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
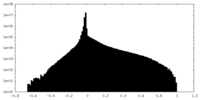
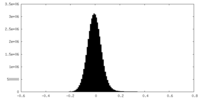
| Density Histograms |
-Half map: Half Map B
| File | emd_28929_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
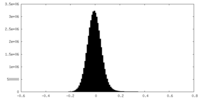
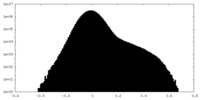
| Density Histograms |
- Sample components
Sample components
-Entire : KWOCA51
| Entire | Name: KWOCA51 |
|---|---|
| Components |
|
-Supramolecule #1: KWOCA51
| Supramolecule | Name: KWOCA51 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
-Macromolecule #1: KWOCA51
| Macromolecule | Name: KWOCA51 / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Sequence | String: METDTLLLWV LLLWVPGSTG DGSHHHHHHG GSEQKLISEE DLSGGGSWSG SSDEEEAREW AERAEEAAKE ALEQAKREGD EIARLCAKML EILAEEARRK KDSEEAEAVY WAARAVLAAL EALEQAKREG DEDARRCAEE LLRLACSAAA RQDSEQARAV YEAARAVLAA ...String: METDTLLLWV LLLWVPGSTG DGSHHHHHHG GSEQKLISEE DLSGGGSWSG SSDEEEAREW AERAEEAAKE ALEQAKREGD EIARLCAKML EILAEEARRK KDSEEAEAVY WAARAVLAAL EALEQAKREG DEDARRCAEE LLRLACSAAA RQDSEQARAV YEAARAVLAA LRALEAAKRA GMEEARKEAE ELLRRACEAA RKQDPELARA VRDKAELLKA LADLFKALKE LKKSLDELER SLEELEKNPS EDALVENNRL NVENNKIIVE VLRIIAEVLR INARAV |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 3.0 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 65.476 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: -1.8 µm / Nominal defocus min: -0.7000000000000001 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: OTHER / Details: Ab initio 3D - cryosparc |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 5.87 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 397630 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)