-Search query
-Search result
Showing all 47 items for (author: gagnon & mg)

EMDB-29819: 
Structure of the Escherichia coli 70S ribosome in complex with EF-Tu and Ile-tRNAIle(LAU) bound to the cognate AUA codon (Structure I)
Method: single particle / : Rybak MY, Gagnon MG

EMDB-29820: 
Structure of the Escherichia coli 70S ribosome in complex with EF-Tu and Ile-tRNAIle(LAU) bound to the near-cognate AUG codon (Structure II)
Method: single particle / : Rybak MY, Gagnon MG

EMDB-29821: 
Structure of the Escherichia coli 70S ribosome in complex with A-site tRNAIle(LAU) bound to the cognate AUA codon (Structure III)
Method: single particle / : Rybak MY, Gagnon MG

EMDB-29822: 
Structure of the Escherichia coli 70S ribosome in complex with P-site tRNAIle(LAU) bound to the cognate AUA codon (Structure IV)
Method: single particle / : Rybak MY, Gagnon MG

EMDB-42554: 
Cryo-EM structure of the Listeria innocua 70S ribosome in complex with HPF (structure I-A)
Method: single particle / : Seely SM, Basu RS, Gagnon MG

EMDB-42555: 
Refined map of the Listeria innocua 70S ribosome (head-swiveled) in complex with pe/E-tRNA (structure I-B)
Method: single particle / : Seely SM, Basu R, Gagnon MG

EMDB-42556: 
Focus refined map of the swiveled head domain of the 30S subunit of Listeria innocua ribosome (structure I-B)
Method: single particle / : Seely SM, Basu RS, Gagnon MG

EMDB-42557: 
Cryo-EM structure of the Listeria innocua 70S ribosome (head-swiveled) in complex with pe/E-tRNA (structure I-B)
Method: single particle / : Seely SM, Basu RS, Gagnon MG

EMDB-42558: 
Refined map of the ratcheted Listeria innocua 70S ribosome in complex with p/E-tRNA (structure II-A)
Method: single particle / : Seely SM, Basu RS, Gagnon MG

EMDB-42559: 
Focus refined map of the head domain of the ratcheted 30S subunit of Listeria innocua ribosome (structure II-A)
Method: single particle / : Seely SM, Basu RS, Gagnon MG

EMDB-42560: 
Focus refined map of the body domain of the ratcheted 30S subunit of Listeria innocua ribosome (structure II-A)
Method: single particle / : Seely SM, Basu RS, Gagnon MG

EMDB-42561: 
Cryo-EM structure of the ratcheted Listeria innocua 70S ribosome in complex with p/E-tRNA (structure II-A)
Method: single particle / : Seely SM, Basu RS, Gagnon MG

EMDB-42562: 
Refined map of the Listeria innocua 70S ribosome in complex with HflXr, HPF, and E-site tRNA (structure II-B)
Method: single particle / : Seely SM, Basu RS, Gagnon MG

EMDB-42563: 
Focus refined map of the head domain of the 30S subunit of Listeria innocua ribosome in complex with HflXr, HPF, and E-site tRNA (structure II-B)
Method: single particle / : Seely SM, Basu RS, Gagnon MG

EMDB-42564: 
Focus refined map of the body domain of the 30S subunit of Listeria innocua ribosome in complex with HflXr, HPF, and E-site tRNA (structure II-B)
Method: single particle / : Seely SM, Basu RS, Gagnon MG

EMDB-42565: 
Focus refined map of HflXr bound to the Listeria innocua ribosome in complex with HPF and E-site tRNA (structure II-B)
Method: single particle / : Seely SM, Basu RS, Gagnon MG

EMDB-42566: 
Cryo-EM structure of the Listeria innocua 70S ribosome in complex with HflXr, HPF, and E-site tRNA (structure II-B)
Method: single particle / : Seely SM, Basu RS, Gagnon MG

EMDB-42567: 
Refined map of the Listeria innocua 70S ribosome (head-swiveled) in complex with HflXr and pe/E-tRNA (structure II-C)
Method: single particle / : Seely SM, Basu RS, Gagnon MG

EMDB-42568: 
Focus refined map of the swiveled head domain of the 30S subunit of Listeria innocua ribosome in complex with HflXr and pe/E-tRNA (structure II-C)
Method: single particle / : Seely SM, Basu RS, Gagnon MG

EMDB-42569: 
Focus refined map of the body domain of the 30S subunit of Listeria innocua ribosome in complex with HflXr and pe/E-tRNA (structure II-C)
Method: single particle / : Seely SM, Basu RS, Gagnon MG

EMDB-42570: 
Focus refined map of HflXr bound to the Listeria innocua ribosome in complex with pe/E-tRNA (structure II-C)
Method: single particle / : Seely SM, Basu RS, Gagnon MG

EMDB-42571: 
Cryo-EM structure of the Listeria innocua 70S ribosome (head-swiveled) in complex with HflXr and pe/E-tRNA (structure II-C)
Method: single particle / : Seely SM, Basu RS, Gagnon MG

EMDB-42572: 
Refined map of the ratcheted Listeria innocua 70S ribosome (head-swiveled) in complex with HflXr and pe/E-tRNA (structure II-D)
Method: single particle / : Seely SM, Basu RS, Gagnon MG

EMDB-42573: 
Focus refined map of the swiveled head domain of the 30S subunit of ratcheted Listeria innocua ribosome in complex with HflXr and pe/E-tRNA (structure II-D)
Method: single particle / : Seely SM, Basu RS, Gagnon MG

EMDB-42574: 
Focus refined map of the body domain of the 30S subunit of ratcheted Listeria innocua ribosome in complex with HflXr and pe/E-tRNA (structure II-D)
Method: single particle / : Seely SM, Basu RS, Gagnon MG

EMDB-42575: 
Focus refined map of HflXr bound to the ratcheted Listeria innocua ribosome in complex with pe/E-tRNA (structure II-D)
Method: single particle / : Seely SM, Basu RS, Gagnon MG

EMDB-42576: 
Cryo-EM structure of the ratcheted Listeria innocua 70S ribosome (head-swiveled) in complex with HflXr and pe/E-tRNA (structure II-D)
Method: single particle / : Seely SM, Basu RS, Gagnon MG

EMDB-42577: 
Cryo-EM structure of the Listeria innocua 50S ribosomal subunit in complex with HflXr (structure III)
Method: single particle / : Seely SM, Basu RS, Gagnon MG

EMDB-19067: 
Cryo-EM structure of P. urativorans 70S ribosome in complex with hibernation factors Balon and RaiA (structure 1).
Method: single particle / : Helena-Bueno K, Rybak MY, Gagnon MG, Hill CH, Melnikov SV

EMDB-19076: 
Cryo-EM structure of P. urativorans 70S ribosome in complex with hibernation factor Balon, mRNA and P-site tRNA (structure 2).
Method: single particle / : Helena-Bueno K, Rybak MY, Gagnon MG, Hill CH, Melnikov SV

EMDB-19077: 
Cryo-EM structure of P. urativorans 70S ribosome in complex with hibernation factor Balon and EF-Tu(GDP) (structure 3).
Method: single particle / : Helena-Bueno K, Rybak MY, Gagnon MG, Hill CH, Melnikov SV
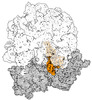
EMDB-43074: 
Cryo-EM structure of the Mycobacterium smegmatis 70S ribosome in complex with hibernation factor Msmeg1130 (Balon) (Structure 4)
Method: single particle / : Rybak MY, Helena-Bueno K, Hill CH, Melnikov SV, Gagnon MG
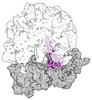
EMDB-43075: 
Cryo-EM structure of the Mycobacterium smegmatis 70S ribosome in complex with hibernation factor Rv2629 (Balon) (Structure 5)
Method: single particle / : Rybak MY, Helena-Bueno K, Hill CH, Melnikov SV, Gagnon MG
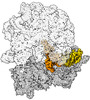
EMDB-43076: 
Cryo-EM structure of the Mycobacterium smegmatis 70S ribosome in complex with hibernation factor Msmeg1130 (Balon) and MsmegEF-Tu(GDP) (Composite structure 6)
Method: single particle / : Rybak MY, Helena-Bueno K, Hill CH, Melnikov SV, Gagnon MG

EMDB-43077: 
Cryo-EM structure of the Mycobacterium smegmatis 70S ribosome in complex with hibernation factor Msmeg1130 (Balon) and MsmegEF-Tu(GDP) (Structure 6)
Method: single particle / : Rybak MY, Helena-Bueno K, Hill CH, Melnikov SV, Gagnon MG

EMDB-43078: 
Hibernation factor Msmeg1130 (Balon) and MsmegEF-Tu(GDP) bound to Mycobacterium smegmatis 70S ribosome, from focused 3D classification and refinement (Structure 6)
Method: single particle / : Rybak MY, Helena-Bueno K, Hill CH, Melnikov SV, Gagnon MG
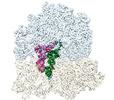
EMDB-40882: 
Cryo-EM structure of the Escherichia coli 70S ribosome in complex with amikacin, mRNA, and A-, P-, and E-site tRNAs
Method: single particle / : Seely SM, Gagnon MG

EMDB-28197: 
Escherichia coli 70S ribosome bound to thermorubin, deacylated P-site tRNAfMet and aminoacylated A-site Phe-tRNA
Method: single particle / : Rybak MY, Gagnon MG
EMDB-26553: 
N2 sub-domain of IF2 bound to the 30S subunit in the Pseudomonas aeruginosa 70S ribosome initiation complex (focused classification and refinement)
Method: single particle / : Basu RS, Sherman MB, Gagnon MG

EMDB-26628: 
Pseudomonas aeruginosa 70S ribosome initiation complex bound to compact IF2-GDP (I-A)
Method: single particle / : Basu RS, Sherman MB, Gagnon MG

EMDB-26629: 
Compact IF2-GDP bound to the Pseudomonas aeruginosa 70S ribosome initiation complex, from focused classification and refinement (I-A)
Method: single particle / : Basu RS, Sherman MB, Gagnon MG

EMDB-26630: 
Pseudomonas aeruginosa 70S ribosome initiation complex bound to compact IF2-GDP (composite structure I-A)
Method: single particle / : Basu RS, Sherman MB, Gagnon MG

EMDB-26631: 
Pseudomonas aeruginosa 70S ribosome initiation complex bound to compact IF2-GDP (I-B)
Method: single particle / : Basu RS, Sherman MB, Gagnon MG

EMDB-26632: 
Compact IF2-GDP bound to the Pseudomonas aeruginosa 70S ribosome initiation complex, from focused classification and refinement (I-B)
Method: single particle / : Basu RS, Sherman MB, Gagnon MG

EMDB-26633: 
Pseudomonas aeruginosa 70S ribosome initiation complex bound to compact IF2-GDP (composite structure I-B)
Method: single particle / : Basu RS, Sherman MB, Gagnon MG

EMDB-26634: 
Pseudomonas aeruginosa 70S ribosome initiation complex bound to IF2-GDPCP (structure II-A)
Method: single particle / : Basu RS, Sherman MB, Gagnon MG

EMDB-26635: 
Pseudomonas aeruginosa 70S ribosome initiation complex bound to IF2-GDPCP (structure II-B)
Method: single particle / : Basu RS, Sherman MB, Gagnon MG
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
