[English] 日本語
 Yorodumi
Yorodumi- EMDB-26629: Compact IF2-GDP bound to the Pseudomonas aeruginosa 70S ribosome ... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Compact IF2-GDP bound to the Pseudomonas aeruginosa 70S ribosome initiation complex, from focused classification and refinement (I-A) | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Initiation Factor 2 / 70S ribosome / cryo-EM / translation initiation / initiator tRNA / conformational changes / RIBOSOME | |||||||||
| Function / homology |  Function and homology information Function and homology informationtranslation initiation factor activity / translational initiation / GTPase activity / GTP binding / cytoplasm / cytosol Similarity search - Function | |||||||||
| Biological species |  Pseudomonas aeruginosa PAO1 (bacteria) Pseudomonas aeruginosa PAO1 (bacteria) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.4 Å | |||||||||
 Authors Authors | Basu RS / Sherman MB / Gagnon MG | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: Compact IF2 allows initiator tRNA accommodation into the P site and gates the ribosome to elongation Authors: Basu RS / Sherman MB / Gagnon MG | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_26629.map.gz emd_26629.map.gz | 255.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-26629-v30.xml emd-26629-v30.xml emd-26629.xml emd-26629.xml | 19.6 KB 19.6 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_26629.png emd_26629.png | 193.1 KB | ||
| Masks |  emd_26629_msk_1.map emd_26629_msk_1.map | 512 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-26629.cif.gz emd-26629.cif.gz | 6.4 KB | ||
| Others |  emd_26629_additional_1.map.gz emd_26629_additional_1.map.gz emd_26629_half_map_1.map.gz emd_26629_half_map_1.map.gz emd_26629_half_map_2.map.gz emd_26629_half_map_2.map.gz | 418.7 MB 474.4 MB 474.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-26629 http://ftp.pdbj.org/pub/emdb/structures/EMD-26629 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26629 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26629 | HTTPS FTP |
-Related structure data
| Related structure data |  7unqMC  7unrC  7untC  7unuC  7unvC  7unwC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
| EM raw data |  EMPIAR-11012 (Title: Pseudomonas aeruginosa 70S ribosome initiation complex bound to compact IF2 EMPIAR-11012 (Title: Pseudomonas aeruginosa 70S ribosome initiation complex bound to compact IF2Data size: 1.7 TB Data #1: Unaligned multiframe micrographs of the Pseudomonas aeruginosa 70S ribosome initiation complex bound to IF2 collected on Gatan K3 in Super Resolution [micrographs - multiframe]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_26629.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_26629.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.85 Å | ||||||||||||||||||||||||||||||||||||
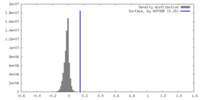
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_26629_msk_1.map emd_26629_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
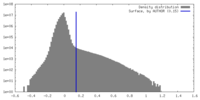
| Density Histograms |
-Additional map: Compact IF2-GDP bound to the Pseudomonas aeruginosa 70S...
| File | emd_26629_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Compact IF2-GDP bound to the Pseudomonas aeruginosa 70S ribosome initiation complex (I-A). Sharpened map. | ||||||||||||
| Projections & Slices |
| ||||||||||||
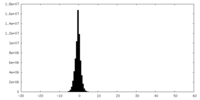
| Density Histograms |
-Half map: Compact IF2-GDP bound to the Pseudomonas aeruginosa 70S...
| File | emd_26629_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Compact IF2-GDP bound to the Pseudomonas aeruginosa 70S ribosome initiation complex (I-A). Half-map. | ||||||||||||
| Projections & Slices |
| ||||||||||||
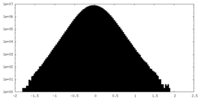
| Density Histograms |
-Half map: Compact IF2-GDP bound to the Pseudomonas aeruginosa 70S...
| File | emd_26629_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Compact IF2-GDP bound to the Pseudomonas aeruginosa 70S ribosome initiation complex (I-A). Half-map. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Compact IF2-GDP bound to the Pseudomonas aeruginosa 70S ribosome ...
| Entire | Name: Compact IF2-GDP bound to the Pseudomonas aeruginosa 70S ribosome initiation complex, from focused classification and refinement (I-A) |
|---|---|
| Components |
|
-Supramolecule #1: Compact IF2-GDP bound to the Pseudomonas aeruginosa 70S ribosome ...
| Supramolecule | Name: Compact IF2-GDP bound to the Pseudomonas aeruginosa 70S ribosome initiation complex, from focused classification and refinement (I-A) type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Pseudomonas aeruginosa PAO1 (bacteria) Pseudomonas aeruginosa PAO1 (bacteria) |
-Macromolecule #1: Translation initiation factor IF-2
| Macromolecule | Name: Translation initiation factor IF-2 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Pseudomonas aeruginosa PAO1 (bacteria) Pseudomonas aeruginosa PAO1 (bacteria) |
| Molecular weight | Theoretical: 91.049641 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MTQVTVKELA QVVDTPVERL LLQMRDAGLP HTSAEQVVTD SEKQALLTHL KGSHGDRASE PRKITLQRKT TTTLKVGGSK TVSVEVRKK KTYVKRSPDE IEAERQRELE EQRAAEEAER LKAEEAAARQ RAEEEARKAE EAARAKAAQE AAATAGAEPA V VADVAVAE ...String: MTQVTVKELA QVVDTPVERL LLQMRDAGLP HTSAEQVVTD SEKQALLTHL KGSHGDRASE PRKITLQRKT TTTLKVGGSK TVSVEVRKK KTYVKRSPDE IEAERQRELE EQRAAEEAER LKAEEAAARQ RAEEEARKAE EAARAKAAQE AAATAGAEPA V VADVAVAE PVAKPAAVEE RKKEEPRRVP KRDEDDDRRD RKHTQHRPSV KEKEKVPAPR VAPRSTDEES DGYRRGGRGG KS KLKKRNQ HGFQNPTGPI VREVNIGETI TVAELAAQMS VKGAEVVKFM FKMGSPVTIN QVLDQETAQL VAEELGHKVK LVS ENALEE QLAESLKFEG EAVTRAPVVT VMGHVDHGKT SLLDYIRRAK VAAGEAGGIT QHIGAYHVET ERGMVTFLDT PGHA AFTAM RARGAQATDI VILVVAADDG VMPQTQEAVQ HAKAAGVPIV VAVNKIDKPE ANPDNIKNGL AALDVIPEEW GGDAP FVPV SAKLGTGVDE LLEAVLLQAE VLELKATPSA PGRGVVVESR LDKGRGPVAT VLVQDGTLRQ GDMVLVGINY GRVRAM LDE NGKPIKEAGP SIPVEILGLD GTPDAGDEMT VVADEKKARE VALFRQGKFR EVKLARAHAG KLENIFENMG QEEKKTL NI VLKADVRGSL EALQGSLSGL GNDEVQVRVV GGGVGGITES DANLALASNA VLFGFNVRAD AGARKIVEAE GLDMRYYN V IYDIIEDVKK ALTGMLGSDL RENILGIAEV RDVFRSPKFG AIAGCMVTEG MVHRNRPIRV LRDDVVIFEG ELESLRRFK DDVAEVRAGM ECGIGVKSYN DVKVGDKIEV FEKVEVARSL UniProtKB: Translation initiation factor IF-2 |
-Macromolecule #2: GUANOSINE-5'-DIPHOSPHATE
| Macromolecule | Name: GUANOSINE-5'-DIPHOSPHATE / type: ligand / ID: 2 / Number of copies: 1 / Formula: GDP |
|---|---|
| Molecular weight | Theoretical: 443.201 Da |
| Chemical component information |  ChemComp-GDP: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.6 |
|---|---|
| Grid | Model: Quantifoil R2/1 / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 85 % / Chamber temperature: 295 K / Instrument: LEICA EM GP |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Number grids imaged: 1 / Number real images: 8056 / Average exposure time: 1.0 sec. / Average electron dose: 31.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.1 µm / Nominal defocus min: 0.2 µm / Nominal magnification: 105000 |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Source name: PDB / Chain - Initial model type: experimental model |
|---|---|
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
| Output model |  PDB-7unq: |
 Movie
Movie Controller
Controller

















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)