[English] 日本語
 Yorodumi
Yorodumi- EMDB-42577: Cryo-EM structure of the Listeria innocua 50S ribosomal subunit i... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the Listeria innocua 50S ribosomal subunit in complex with HflXr (structure III) | |||||||||||||||
 Map data Map data | Cryo-EM map of the Listeria innocua 50S ribosomal subunit in complex with HflXr (structure III) | |||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | cryo-EM / recycling / HflXr / time-resolved / RIBOSOME | |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationlarge ribosomal subunit / transferase activity / ribosome binding / 5S rRNA binding / ribosomal large subunit assembly / large ribosomal subunit rRNA binding / cytosolic large ribosomal subunit / cytoplasmic translation / tRNA binding / negative regulation of translation ...large ribosomal subunit / transferase activity / ribosome binding / 5S rRNA binding / ribosomal large subunit assembly / large ribosomal subunit rRNA binding / cytosolic large ribosomal subunit / cytoplasmic translation / tRNA binding / negative regulation of translation / rRNA binding / structural constituent of ribosome / ribosome / translation / ribonucleoprotein complex / mRNA binding / GTPase activity / GTP binding / metal ion binding / cytoplasm Similarity search - Function | |||||||||||||||
| Biological species |  Listeria innocua (bacteria) / Listeria innocua (bacteria) /  Listeria monocytogenes serovar 1/2a (strain ATCC BAA-679 / EGD-e) (bacteria) Listeria monocytogenes serovar 1/2a (strain ATCC BAA-679 / EGD-e) (bacteria) | |||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.7 Å | |||||||||||||||
 Authors Authors | Seely SM / Basu RS / Gagnon MG | |||||||||||||||
| Funding support |  United States, 4 items United States, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Nucleic Acids Res / Year: 2024 Journal: Nucleic Acids Res / Year: 2024Title: Mechanistic insights into the alternative ribosome recycling by HflXr. Authors: Savannah M Seely / Ritwika S Basu / Matthieu G Gagnon /  Abstract: During stress conditions such as heat shock and antibiotic exposure, ribosomes stall on messenger RNAs, leading to inhibition of protein synthesis. To remobilize ribosomes, bacteria use rescue ...During stress conditions such as heat shock and antibiotic exposure, ribosomes stall on messenger RNAs, leading to inhibition of protein synthesis. To remobilize ribosomes, bacteria use rescue factors such as HflXr, a homolog of the conserved housekeeping GTPase HflX that catalyzes the dissociation of translationally inactive ribosomes into individual subunits. Here we use time-resolved cryo-electron microscopy to elucidate the mechanism of ribosome recycling by Listeria monocytogenes HflXr. Within the 70S ribosome, HflXr displaces helix H69 of the 50S subunit and induces long-range movements of the platform domain of the 30S subunit, disrupting inter-subunit bridges B2b, B2c, B4, B7a and B7b. Our findings unveil a unique ribosome recycling strategy by HflXr which is distinct from that mediated by RRF and EF-G. The resemblance between HflXr and housekeeping HflX suggests that the alternative ribosome recycling mechanism reported here is universal in the prokaryotic kingdom. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_42577.map.gz emd_42577.map.gz | 256.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-42577-v30.xml emd-42577-v30.xml emd-42577.xml emd-42577.xml | 53.5 KB 53.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_42577.png emd_42577.png | 90.6 KB | ||
| Masks |  emd_42577_msk_1.map emd_42577_msk_1.map | 512 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-42577.cif.gz emd-42577.cif.gz | 11.1 KB | ||
| Others |  emd_42577_additional_1.map.gz emd_42577_additional_1.map.gz emd_42577_half_map_1.map.gz emd_42577_half_map_1.map.gz emd_42577_half_map_2.map.gz emd_42577_half_map_2.map.gz | 264.5 MB 475.4 MB 475.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-42577 http://ftp.pdbj.org/pub/emdb/structures/EMD-42577 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42577 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42577 | HTTPS FTP |
-Validation report
| Summary document |  emd_42577_validation.pdf.gz emd_42577_validation.pdf.gz | 952.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_42577_full_validation.pdf.gz emd_42577_full_validation.pdf.gz | 952.4 KB | Display | |
| Data in XML |  emd_42577_validation.xml.gz emd_42577_validation.xml.gz | 19.1 KB | Display | |
| Data in CIF |  emd_42577_validation.cif.gz emd_42577_validation.cif.gz | 22.8 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-42577 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-42577 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-42577 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-42577 | HTTPS FTP |
-Related structure data
| Related structure data |  8uuaMC  8uu4C  8uu5C  8uu6C  8uu7C  8uu8C  8uu9C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_42577.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_42577.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM map of the Listeria innocua 50S ribosomal subunit in complex with HflXr (structure III) | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.85 Å | ||||||||||||||||||||||||||||||||||||
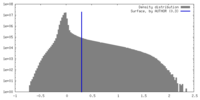
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_42577_msk_1.map emd_42577_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Listeria innocua 50S ribosomal subunit in complex with...
| File | emd_42577_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Listeria innocua 50S ribosomal subunit in complex with HflXr (structure III) Sharpened map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Listeria innocua 50S ribosomal subunit in complex with...
| File | emd_42577_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Listeria innocua 50S ribosomal subunit in complex with HflXr (structure III) Half map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Listeria innocua 50S ribosomal subunit in complex with...
| File | emd_42577_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Listeria innocua 50S ribosomal subunit in complex with HflXr (structure III) Half map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
+Entire : Listeria innocua 50S ribosomal subunit bound to HflXr
+Supramolecule #1: Listeria innocua 50S ribosomal subunit bound to HflXr
+Macromolecule #1: GTPase HflX
+Macromolecule #4: Large ribosomal subunit protein uL2
+Macromolecule #5: Large ribosomal subunit protein uL3
+Macromolecule #6: Large ribosomal subunit protein uL4
+Macromolecule #7: Large ribosomal subunit protein uL5
+Macromolecule #8: Large ribosomal subunit protein uL6
+Macromolecule #9: Large ribosomal subunit protein uL13
+Macromolecule #10: Large ribosomal subunit protein uL14
+Macromolecule #11: Large ribosomal subunit protein uL15
+Macromolecule #12: Large ribosomal subunit protein uL16
+Macromolecule #13: Large ribosomal subunit protein bL17
+Macromolecule #14: Large ribosomal subunit protein uL18
+Macromolecule #15: Large ribosomal subunit protein bL19
+Macromolecule #16: Large ribosomal subunit protein bL20
+Macromolecule #17: Large ribosomal subunit protein bL21
+Macromolecule #18: Large ribosomal subunit protein uL22
+Macromolecule #19: Large ribosomal subunit protein uL23
+Macromolecule #20: Large ribosomal subunit protein uL24
+Macromolecule #21: Large ribosomal subunit protein bL27
+Macromolecule #22: Large ribosomal subunit protein bL28
+Macromolecule #23: Large ribosomal subunit protein uL29
+Macromolecule #24: Large ribosomal subunit protein uL30
+Macromolecule #25: Large ribosomal subunit protein bL31B
+Macromolecule #26: Large ribosomal subunit protein bL32
+Macromolecule #27: Large ribosomal subunit protein bL33
+Macromolecule #28: Large ribosomal subunit protein bL34
+Macromolecule #29: Large ribosomal subunit protein bL35
+Macromolecule #30: Large ribosomal subunit protein bL36
+Macromolecule #2: 23S Ribosomal RNA
+Macromolecule #3: 5S Ribosomal RNA
+Macromolecule #31: PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER
+Macromolecule #32: MAGNESIUM ION
+Macromolecule #33: POTASSIUM ION
+Macromolecule #34: ZINC ION
+Macromolecule #35: water
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.6 |
|---|---|
| Grid | Model: Quantifoil R2/1 / Material: GOLD / Mesh: 200 / Support film - Material: GOLD / Support film - topology: HOLEY / Pretreatment - Type: PLASMA CLEANING / Pretreatment - Time: 30 sec. / Pretreatment - Atmosphere: OTHER |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 85 % / Chamber temperature: 295 K / Instrument: LEICA EM GP |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: INTEGRATING / Number grids imaged: 1 / Number real images: 10446 / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.3000000000000003 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 96000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)