-Search query
-Search result
Showing 1 - 50 of 432 items for (author: chand & d)

EMDB-41036: 
Closed human HCN1 F186C S264C bound to cAMP, reconstituted in LMNG + SPL
Method: single particle / : Burtscher V, Mount J, Cowgill J, Chang Y, Bickel K, Yuan P, Chanda B

PDB-8t4m: 
Closed human HCN1 F186C S264C bound to cAMP, reconstituted in LMNG + SPL
Method: single particle / : Burtscher V, Mount J, Cowgill J, Chang Y, Bickel K, Yuan P, Chanda B

EMDB-29950: 
SARS-Cov2 S protein structure in complex with neutralizing monoclonal antibody 002-S21B10
Method: single particle / : Patel A, Ortlund EA

EMDB-29975: 
Overall map of SARS-Cov2 S protein structure in complex with neutralizing monoclonal antibody 002-S21B10
Method: single particle / : Patel A, Ortlund EA

EMDB-40007: 
Local map of SARS-Cov2 S protein structure in complex with neutralizing monoclonal antibody 002-S21B10
Method: single particle / : Patel A, Ortlund EA

EMDB-41711: 
Disulfide-stabilized HIV-1 CA hexamer in complex with PQBP1 Nt
Method: single particle / : Piacentini J, Pornillos O, Ganser-Pornillos BK

PDB-8ty6: 
Disulfide-stabilized HIV-1 CA hexamer in complex with PQBP1 Nt
Method: single particle / : Piacentini J, Pornillos O, Ganser-Pornillos BK
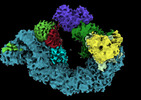
EMDB-19567: 
Closed crosslinked structure of (NEDD8)-CRL2VHL-MZ1-Brd4BD2-Ub(G76S, K48C)-UBE2R1(C93K, S138C, C191S, C223S)-Ub
Method: single particle / : Ciulli A, Crowe C, Nakasone MA

EMDB-40967: 
Cryo-EM structure of a full-length, native Drp1 dimer
Method: single particle / : Rochon K, Mears JA

PDB-8t1h: 
Cryo-EM structure of a full-length, native Drp1 dimer
Method: single particle / : Rochon K, Mears JA
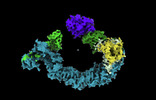
EMDB-19569: 
Open non-crosslinked structure Brd4BD2-MZ1-(NEDD8)-CRL2VHL
Method: single particle / : Ciulli A, Crowe C, Nakasone MA

EMDB-40699: 
SARS-CoV-2 replication-transcription complex bound to nsp9 and UMPCPP, as a pre-catalytic NMPylation intermediate
Method: single particle / : Small GI, Darst SA, Campbell EA

EMDB-40707: 
SARS-CoV-2 replication-transcription complex bound to RNA-nsp9, as a noncatalytic RNA-nsp9 binding mode
Method: single particle / : Small GI, Darst SA, Campbell EA

EMDB-40708: 
SARS-CoV-2 replication-transcription complex bound to RNA-nsp9 and GDP-betaS, as a pre-catalytic deRNAylation/mRNA capping intermediate
Method: single particle / : Small GI, Darst SA, Campbell EA

PDB-8sq9: 
SARS-CoV-2 replication-transcription complex bound to nsp9 and UMPCPP, as a pre-catalytic NMPylation intermediate
Method: single particle / : Small GI, Darst SA, Campbell EA

PDB-8sqj: 
SARS-CoV-2 replication-transcription complex bound to RNA-nsp9, as a noncatalytic RNA-nsp9 binding mode
Method: single particle / : Small GI, Darst SA, Campbell EA

PDB-8sqk: 
SARS-CoV-2 replication-transcription complex bound to RNA-nsp9 and GDP-betaS, as a pre-catalytic deRNAylation/mRNA capping intermediate
Method: single particle / : Small GI, Darst SA, Campbell EA

EMDB-41075: 
SARS-CoV-2 spike in complex with Fab 71281-33
Method: single particle / : Binshtein E, Crowe JE

EMDB-41076: 
SARS-CoV-2 spike in complex with Fab 71281-33 (2)
Method: single particle / : Binshtein E, Crowe JE

EMDB-41153: 
Integrin alpha-v beta-8 in complex with minibinder B8_BP_dsulf
Method: single particle / : Campbell MG, Fernandez A, Roy A, Kraft J, Baker D

EMDB-41154: 
Integrin alpha-v beta-6 in complex with minibinder B6_BP_dslf
Method: single particle / : Campbell MG, Fernandez A, Roy A, Kraft J, Baker D

PDB-8tcf: 
Integrin alpha-v beta-8 in complex with minibinder B8_BP_dsulf
Method: single particle / : Campbell MG, Fernandez A, Roy A, Kraft J, Baker D

PDB-8tcg: 
Integrin alpha-v beta-6 in complex with minibinder B6_BP_dslf
Method: single particle / : Campbell MG, Fernandez A, Roy A, Kraft J, Baker D

EMDB-16052: 
Cryo-EM structure of stalled rabbit 80S ribosomes in complex with human CCR4-NOT and CNOT4
Method: single particle / : Absmeier E, Chandrasekaran V, O'Reilly FJ, Stowell JAW, Rappsilber J, Passmore LA

PDB-8bhf: 
Cryo-EM structure of stalled rabbit 80S ribosomes in complex with human CCR4-NOT and CNOT4
Method: single particle / : Absmeier E, Chandrasekaran V, O'Reilly FJ, Stowell JAW, Rappsilber J, Passmore LA

EMDB-29077: 
Cryo-EM structure of the STAR-0215 Fab in complex with active human plasma kallikrein
Method: single particle / : Fuller JR, Biris N, Bista P

PDB-8fgx: 
Cryo-EM structure of the STAR-0215 Fab in complex with active human plasma kallikrein
Method: single particle / : Fuller JR, Biris N, Bista P

EMDB-27808: 
RSV F trimer bound to RSV-199 Fab
Method: single particle / : Wen X, Jardetzky TS

EMDB-27846: 
HMPV F monomer bound to RSV-199 Fab
Method: single particle / : Wen X, Jardetzky TS

EMDB-27990: 
HMPV F complex with 4I3 Fab
Method: single particle / : Wen X, Jardetzky TS

EMDB-27995: 
HMPV F dimer bound to RSV-199 Fab
Method: single particle / : Wen X, Jardetzky TS

PDB-8dzw: 
RSV F trimer bound to RSV-199 Fab
Method: single particle / : Wen X, Jardetzky TS

PDB-8e2u: 
HMPV F monomer bound to RSV-199 Fab
Method: single particle / : Wen X, Jardetzky TS

PDB-8eay: 
HMPV F complex with 4I3 Fab
Method: single particle / : Wen X, Jardetzky TS

PDB-8ebp: 
HMPV F dimer bound to RSV-199 Fab
Method: single particle / : Wen X, Jardetzky TS

EMDB-34551: 
Cryo-EM structure of HACE1 monomer
Method: single particle / : Singh S, Machida S, Tulsian NK, Choong YK, Ng J, Shanker S, Yaochen LD, Shi J, Sivaraman J

EMDB-34586: 
Cryo-EM structure of HACE1 dimer
Method: single particle / : Singh S, Machida S, Tulsian NK, Choong YK, Ng J, Shanker S, Yaochen LD, Shi J, Sivaraman J

PDB-8h8x: 
Cryo-EM structure of HACE1 monomer
Method: single particle / : Singh S, Machida S, Tulsian NK, Choong YK, Ng J, Shanker S, Yaochen LD, Shi J, Sivaraman J

PDB-8hae: 
Cryo-EM structure of HACE1 dimer
Method: single particle / : Singh S, Machida S, Tulsian NK, Choong YK, Ng J, Shanker S, Yaochen LD, Shi J, Sivaraman J, Machida S

EMDB-29857: 
Molecular mechanism of nucleotide inhibition of human uncoupling protein 1
Method: single particle / : Gogoi P, Jones SA, Ruprecht JJ, King MS, Lee Y, Zogg T, Pardon E, Chand D, Steimle S, Copeman D, Cotrim CA, Steyaert J, Crichton PG, Moiseenkova-Bell V, Kunji ERS

PDB-8g8w: 
Molecular mechanism of nucleotide inhibition of human uncoupling protein 1
Method: single particle / : Gogoi P, Jones SA, Ruprecht JJ, King MS, Lee Y, Zogg T, Pardon E, Chand D, Steimle S, Copeman D, Cotrim CA, Steyaert J, Crichton PG, Moiseenkova-Bell V, Kunji ERS

EMDB-16799: 
Cryo-EM structure of the NINJ1 filament
Method: helical / : Degen MD, Hiller SH, Maier TM
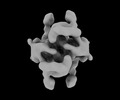
EMDB-16914: 
Cryo-EM structure of the DnaD-NTD tetramer
Method: single particle / : Winterhalter C, Pelliciari S, Cronin N, Costa TRD, Murray H, Ilangovan A

PDB-8ojj: 
Cryo-EM structure of the DnaD-NTD tetramer
Method: single particle / : Winterhalter C, Pelliciari S, Cronin N, Costa TRD, Murray H, Ilangovan A

PDB-8sah: 
Huntingtin C-HEAT domain in complex with HAP40
Method: single particle / : Harding RJ, Deme JC, Alteen MG, Arrowsmith CH, Lea SM, Structural Genomics Consortium (SGC)

EMDB-29790: 
30S focus refined map of WT E.coli ribosome complexed with A-site ortho-aminobenzoic acid charged tRNA-Phe
Method: single particle / : Majumdar C, Cate JHD

EMDB-26656: 
SARS-Cov2 S protein structure in complex with neutralizing monoclonal antibody 034_32
Method: single particle / : Patel A, Ortlund E

PDB-7uow: 
SARS-Cov2 S protein structure in complex with neutralizing monoclonal antibody 034_32
Method: single particle / : Patel A, Ortlund E

EMDB-26562: 
Cryo-EM structure of Human respiratory syncytial virus F variant (construct pXCS847A)
Method: single particle / : Lees JA, Ammirati M, Han S
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search




 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
