[English] 日本語
 Yorodumi
Yorodumi- EMDB-26562: Cryo-EM structure of Human respiratory syncytial virus F variant ... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of Human respiratory syncytial virus F variant (construct pXCS847A) | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | RSV / RSVF / pre-fusion / VIRAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationsymbiont-mediated induction of syncytium formation / host cell Golgi membrane / entry receptor-mediated virion attachment to host cell / fusion of virus membrane with host plasma membrane / viral envelope / symbiont entry into host cell / host cell plasma membrane / virion membrane Similarity search - Function | |||||||||
| Biological species |  Respiratory syncytial virus / Respiratory syncytial virus /  | |||||||||
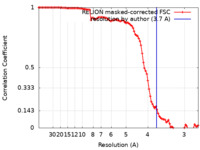
| Method | single particle reconstruction / cryo EM / Resolution: 3.7 Å | |||||||||
 Authors Authors | Lees JA / Ammirati M / Han S | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Sci Transl Med / Year: 2023 Journal: Sci Transl Med / Year: 2023Title: Rational design of a highly immunogenic prefusion-stabilized F glycoprotein antigen for a respiratory syncytial virus vaccine. Authors: Ye Che / Alexey V Gribenko / Xi Song / Luke D Handke / Kari S Efferen / Kristin Tompkins / Srinivas Kodali / Lorna Nunez / A Krishna Prasad / Lynn M Phelan / Mark Ammirati / Xiaodi Yu / ...Authors: Ye Che / Alexey V Gribenko / Xi Song / Luke D Handke / Kari S Efferen / Kristin Tompkins / Srinivas Kodali / Lorna Nunez / A Krishna Prasad / Lynn M Phelan / Mark Ammirati / Xiaodi Yu / Joshua A Lees / Wei Chen / Lyndsey Martinez / Vidia Roopchand / Seungil Han / Xiayang Qiu / John P DeVincenzo / Kathrin U Jansen / Philip R Dormitzer / Kena A Swanson /  Abstract: Respiratory syncytial virus (RSV) is the leading, global cause of serious respiratory disease in infants and is an important cause of respiratory illness in older adults. No RSV vaccine is currently ...Respiratory syncytial virus (RSV) is the leading, global cause of serious respiratory disease in infants and is an important cause of respiratory illness in older adults. No RSV vaccine is currently available. The RSV fusion (F) glycoprotein is a key antigen for vaccine development, and its prefusion conformation is the target of the most potent neutralizing antibodies. Here, we describe a computational and experimental strategy for designing immunogens that enhance the conformational stability and immunogenicity of RSV prefusion F. We obtained an optimized vaccine antigen after screening nearly 400 engineered F constructs. Through in vitro and in vivo characterization studies, we identified F constructs that are more stable in the prefusion conformation and elicit ~10-fold higher serum-neutralizing titers in cotton rats than DS-Cav1. The stabilizing mutations of the lead construct (847) were introduced onto F glycoprotein backbones of strains representing the dominant circulating genotypes of the two major RSV subgroups, A and B. Immunization of cotton rats with a bivalent vaccine formulation of these antigens conferred complete protection against RSV challenge, with no evidence of disease enhancement. The resulting bivalent RSV prefusion F investigational vaccine has recently been shown to be efficacious against RSV disease in two pivotal phase 3 efficacy trials, one for passive protection of infants by immunization of pregnant women and the second for active protection of older adults by direct immunization. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_26562.map.gz emd_26562.map.gz | 78.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-26562-v30.xml emd-26562-v30.xml emd-26562.xml emd-26562.xml | 26.5 KB 26.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_26562_fsc.xml emd_26562_fsc.xml | 9.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_26562.png emd_26562.png | 125.8 KB | ||
| Filedesc metadata |  emd-26562.cif.gz emd-26562.cif.gz | 7.6 KB | ||
| Others |  emd_26562_half_map_1.map.gz emd_26562_half_map_1.map.gz emd_26562_half_map_2.map.gz emd_26562_half_map_2.map.gz | 65.5 MB 65.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-26562 http://ftp.pdbj.org/pub/emdb/structures/EMD-26562 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26562 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26562 | HTTPS FTP |
-Related structure data
| Related structure data |  7ujaMC  7uj3C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_26562.map.gz / Format: CCP4 / Size: 83.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_26562.map.gz / Format: CCP4 / Size: 83.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.37 Å | ||||||||||||||||||||||||||||||||||||
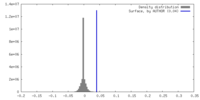
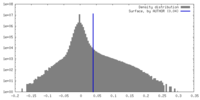
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_26562_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
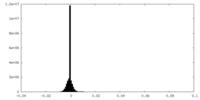
| Density Histograms |
-Half map: #1
| File | emd_26562_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
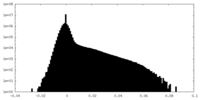
| Density Histograms |
- Sample components
Sample components
-Entire : Human respiratory syncytial virus F variant (construct pXCS847A) ...
| Entire | Name: Human respiratory syncytial virus F variant (construct pXCS847A) with Fabs AM14 and AM22 |
|---|---|
| Components |
|
-Supramolecule #1: Human respiratory syncytial virus F variant (construct pXCS847A) ...
| Supramolecule | Name: Human respiratory syncytial virus F variant (construct pXCS847A) with Fabs AM14 and AM22 type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#6 |
|---|---|
| Source (natural) | Organism:  Respiratory syncytial virus Respiratory syncytial virus |
-Macromolecule #1: RSV variant (construct pXCS847A) F1
| Macromolecule | Name: RSV variant (construct pXCS847A) F1 / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Respiratory syncytial virus Respiratory syncytial virus |
| Molecular weight | Theoretical: 45.025391 KDa |
| Recombinant expression | Organism: Mammalia (mammals) |
| Sequence | String: FLGFLLGVGS ACASGIAVSK VLHLEGEVNK IKSALLSTNK AVVSLSNGVS VLTIKVLDLK NYIDKQLLPI VNKQSCSISN IETVIEFQQ KNNRLLEITR EFSVNAGVTT PVSTYMLTNS ELLSLINDMP ITNDQKKLMS SNVQIVRQQS YSIMSIIKEE V LAYVVQLP ...String: FLGFLLGVGS ACASGIAVSK VLHLEGEVNK IKSALLSTNK AVVSLSNGVS VLTIKVLDLK NYIDKQLLPI VNKQSCSISN IETVIEFQQ KNNRLLEITR EFSVNAGVTT PVSTYMLTNS ELLSLINDMP ITNDQKKLMS SNVQIVRQQS YSIMSIIKEE V LAYVVQLP LYGVIDTPCW KLHTSPLCTT NTKEGSNICL TRTDRGWYCD NAGSVSFFPQ AETCKVQSNR VFCDTMNSLT LP SEVNLCN IDIFNPKYDC KIMTSKTDVS SSVITSLGAI VSCYGKTKCT ASNKNRGIIK TFSNGCDYVS NKGVDTVSVG NTL YYVNKQ EGKSLYVKGE PIINFYDPLV FPSSEFDASI SQVNEKINQS LAFIRKSDEL LSAIGGYIPE APRDGQAYVR KDGE WVLLS TFLGG UniProtKB: Fusion glycoprotein F0 |
-Macromolecule #2: AM14 Fab heavy chain
| Macromolecule | Name: AM14 Fab heavy chain / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 26.231088 KDa |
| Recombinant expression | Organism: Mammalia (mammals) |
| Sequence | String: EVQLVESGGG VVQPGRSLRL SCAASGFSFS HYAMHWVRQA PGKGLEWVAV ISYDGENTYY ADSVKGRFSI SRDNSKNTVS LQMNSLRPE DTALYYCARD RIVDDYYYYG MDVWGQGATV TVSSASTKGP SVFPLAPSSK STSGGTAALG CLVKDYFPEP V TVSWNSGA ...String: EVQLVESGGG VVQPGRSLRL SCAASGFSFS HYAMHWVRQA PGKGLEWVAV ISYDGENTYY ADSVKGRFSI SRDNSKNTVS LQMNSLRPE DTALYYCARD RIVDDYYYYG MDVWGQGATV TVSSASTKGP SVFPLAPSSK STSGGTAALG CLVKDYFPEP V TVSWNSGA LTSGVHTFPA VLQSSGLYSL SSVVTVPSSS LGTQTYICNV NHKPSNTKVD KKVEPKSCDG GGSLVPRGSD YK DDDDK |
-Macromolecule #3: AM14 Fab light chain
| Macromolecule | Name: AM14 Fab light chain / type: protein_or_peptide / ID: 3 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 23.972656 KDa |
| Recombinant expression | Organism: Mammalia (mammals) |
| Sequence | String: DIQMTQSPSS LSASVGDRVT ITCQASQDIK KYLNWYHQKP GKVPELLMHD ASNLETGVPS RFSGRGSGTD FTLTISSLQP EDIGTYYCQ QYDNLPPLTF GGGTKVEIKR TVRTVAAPSV FIFPPSDEQL KSGTASVVCL LNNFYPREAK VQWKVDNALQ S GNSQESVT ...String: DIQMTQSPSS LSASVGDRVT ITCQASQDIK KYLNWYHQKP GKVPELLMHD ASNLETGVPS RFSGRGSGTD FTLTISSLQP EDIGTYYCQ QYDNLPPLTF GGGTKVEIKR TVRTVAAPSV FIFPPSDEQL KSGTASVVCL LNNFYPREAK VQWKVDNALQ S GNSQESVT EQDSKDSTYS LSSTLTLSKA DYEKHKVYAC EVTHQGLSSP VTKSFNRGEC |
-Macromolecule #4: AM22 Fab heavy chain
| Macromolecule | Name: AM22 Fab heavy chain / type: protein_or_peptide / ID: 4 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 25.989154 KDa |
| Recombinant expression | Organism: Mammalia (mammals) |
| Sequence | String: QVQLVQSGAE VKKPGATVKV SCKISGHTLI KLSIHWVRQA PGKGLEWMGG YEGEVDEIFY AQKFQHRLTV IADTATDTVY MELGRLTSD DTAVYFCGTL GVTVTEAGLG IDDYWGQGTL VTVSSASTKG PSVFPLAPSS KSTSGGTAAL GCLVKDYFPE P VTVSWNSG ...String: QVQLVQSGAE VKKPGATVKV SCKISGHTLI KLSIHWVRQA PGKGLEWMGG YEGEVDEIFY AQKFQHRLTV IADTATDTVY MELGRLTSD DTAVYFCGTL GVTVTEAGLG IDDYWGQGTL VTVSSASTKG PSVFPLAPSS KSTSGGTAAL GCLVKDYFPE P VTVSWNSG ALTSGVHTFP AVLQSSGLYS LSSVVTVPSS SLGTQTYICN VNHKPSNTKV DKKVEPKSCD GGGSLVPRGS DY KDDDDK |
-Macromolecule #5: AM22 Fab light chain
| Macromolecule | Name: AM22 Fab light chain / type: protein_or_peptide / ID: 5 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 23.273938 KDa |
| Recombinant expression | Organism: Mammalia (mammals) |
| Sequence | String: EIVLTQSPGT LSLSPGERAT LSCRASQIVS RNHLAWYQQK PGQAPRLLIF GASSRATGIP VRFSGSGSGT DFTLTINGLA PEDFAVYYC LSSDSSIFTF GPGTKVDFKR TVAAPSVFIF PPSDEQLKSG TASVVCLLNN FYPREAKVQW KVDNALQSGN S QESVTEQD ...String: EIVLTQSPGT LSLSPGERAT LSCRASQIVS RNHLAWYQQK PGQAPRLLIF GASSRATGIP VRFSGSGSGT DFTLTINGLA PEDFAVYYC LSSDSSIFTF GPGTKVDFKR TVAAPSVFIF PPSDEQLKSG TASVVCLLNN FYPREAKVQW KVDNALQSGN S QESVTEQD SKDSTYSLSS TLTLSKADYE KHKVYACEVT HQGLSSPVTK SFNRGEC |
-Macromolecule #6: RSV variant (construct pXCS847A) F2
| Macromolecule | Name: RSV variant (construct pXCS847A) F2 / type: protein_or_peptide / ID: 6 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Respiratory syncytial virus Respiratory syncytial virus |
| Molecular weight | Theoretical: 12.109954 KDa |
| Recombinant expression | Organism: Mammalia (mammals) |
| Sequence | String: MELPILKTNA ITTILAAVTL CFASSQNITE EFYQSTCSAV SKGYLSALRT GWYTSVITIE LSNIKENKCN GTDAKVKLIK QELDKYKNA VTELQLLMQS TPACNSRARR UniProtKB: Fusion glycoprotein F0 |
-Macromolecule #7: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 7 / Number of copies: 3 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.1 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| |||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY | |||||||||
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Average exposure time: 20.0 sec. / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 5.0 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 105000 |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)