[English] 日本語
 Yorodumi
Yorodumi- EMDB-19567: Closed crosslinked structure of (NEDD8)-CRL2VHL-MZ1-Brd4BD2-Ub(G7... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Closed crosslinked structure of (NEDD8)-CRL2VHL-MZ1-Brd4BD2-Ub(G76S, K48C)-UBE2R1(C93K, S138C, C191S, C223S)-Ub | ||||||||||||||||||
 Map data Map data | Cryo-EM structure of the (NEDD8)-CRL2VHL-MZ1-Brd4BD2 ternary complex primed for catalysis with UBE2R1-Ub. | ||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||
 Keywords Keywords | E3 ligase / Cullin / PROTAC / BET bromodomain / LIGASE | ||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationregulation of cellular response to hypoxia / negative regulation of beige fat cell differentiation / negative regulation of receptor signaling pathway via JAK-STAT / RHOBTB3 ATPase cycle / cullin-RING-type E3 NEDD8 transferase / (E3-independent) E2 ubiquitin-conjugating enzyme / NEDD8 transferase activity / cullin-RING ubiquitin ligase complex / Cul7-RING ubiquitin ligase complex / cellular response to chemical stress ...regulation of cellular response to hypoxia / negative regulation of beige fat cell differentiation / negative regulation of receptor signaling pathway via JAK-STAT / RHOBTB3 ATPase cycle / cullin-RING-type E3 NEDD8 transferase / (E3-independent) E2 ubiquitin-conjugating enzyme / NEDD8 transferase activity / cullin-RING ubiquitin ligase complex / Cul7-RING ubiquitin ligase complex / cellular response to chemical stress / transcription elongation factor activity / target-directed miRNA degradation / Loss of Function of FBXW7 in Cancer and NOTCH1 Signaling / elongin complex / protein K27-linked ubiquitination / positive regulation of protein autoubiquitination / RNA polymerase II transcription initiation surveillance / protein neddylation / hypothalamus gonadotrophin-releasing hormone neuron development / Replication of the SARS-CoV-1 genome / female meiosis I / NEDD8 ligase activity / positive regulation of protein monoubiquitination / VCB complex / fat pad development / negative regulation of response to oxidative stress / mitochondrion transport along microtubule / Cul5-RING ubiquitin ligase complex / intracellular membraneless organelle / ubiquitin-ubiquitin ligase activity / SCF ubiquitin ligase complex / ubiquitin-dependent protein catabolic process via the C-end degron rule pathway / E2 ubiquitin-conjugating enzyme / Cul2-RING ubiquitin ligase complex / negative regulation of type I interferon production / SCF-dependent proteasomal ubiquitin-dependent protein catabolic process / Cul3-RING ubiquitin ligase complex / female gonad development / SUMOylation of ubiquitinylation proteins / seminiferous tubule development / negative regulation of mitophagy / Cul4A-RING E3 ubiquitin ligase complex / Cul4-RING E3 ubiquitin ligase complex / Prolactin receptor signaling / histone H4K8ac reader activity / Cul4B-RING E3 ubiquitin ligase complex / ubiquitin ligase complex scaffold activity / histone H3K9ac reader activity / RNA polymerase II C-terminal domain binding / histone H3K27ac reader activity / ubiquitin conjugating enzyme activity / TGF-beta receptor signaling activates SMADs / P-TEFb complex binding / male meiosis I / negative regulation of DNA damage checkpoint / histone H4 reader activity / negative regulation of transcription elongation by RNA polymerase II / regulation of proteolysis / Pausing and recovery of Tat-mediated HIV elongation / Tat-mediated HIV elongation arrest and recovery / histone H4K5ac reader activity / cullin family protein binding / histone H4K12ac reader activity / HIV elongation arrest and recovery / Pausing and recovery of HIV elongation / host-mediated suppression of viral transcription / histone H4K16ac reader activity / regulation of postsynapse assembly / positive regulation of intrinsic apoptotic signaling pathway by p53 class mediator / positive regulation of G2/M transition of mitotic cell cycle / DNA replication initiation / protein monoubiquitination / anatomical structure morphogenesis / negative regulation of signal transduction / positive regulation of T-helper 17 cell lineage commitment / Tat-mediated elongation of the HIV-1 transcript / site of DNA damage / Formation of HIV-1 elongation complex containing HIV-1 Tat / ubiquitin-like ligase-substrate adaptor activity / Formation of HIV elongation complex in the absence of HIV Tat / protein K48-linked ubiquitination / energy homeostasis / RNA Polymerase II Transcription Elongation / neuron projection morphogenesis / cellular response to interferon-beta / Formation of RNA Pol II elongation complex / signal transduction in response to DNA damage / Nuclear events stimulated by ALK signaling in cancer / regulation of proteasomal protein catabolic process / negative regulation of TORC1 signaling / transcription-coupled nucleotide-excision repair / Maturation of protein E / Maturation of protein E / RNA Polymerase II Pre-transcription Events / positive regulation of TORC1 signaling / RNA polymerase II CTD heptapeptide repeat kinase activity / regulation of cellular response to insulin stimulus / ER Quality Control Compartment (ERQC) / Myoclonic epilepsy of Lafora / FLT3 signaling by CBL mutants Similarity search - Function | ||||||||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.7 Å | ||||||||||||||||||
 Authors Authors | Ciulli A / Crowe C / Nakasone MA | ||||||||||||||||||
| Funding support | European Union,  United Kingdom, United Kingdom,  Switzerland, 5 items Switzerland, 5 items
| ||||||||||||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2024 Journal: Sci Adv / Year: 2024Title: Mechanism of degrader-targeted protein ubiquitinability. Authors: Charlotte Crowe / Mark A Nakasone / Sarah Chandler / Conner Craigon / Gajanan Sathe / Michael H Tatham / Nikolai Makukhin / Ronald T Hay / Alessio Ciulli /  Abstract: Small-molecule degraders of disease-driving proteins offer a clinically proven modality with enhanced therapeutic efficacy and potential to tackle previously undrugged targets. Stable and long-lived ...Small-molecule degraders of disease-driving proteins offer a clinically proven modality with enhanced therapeutic efficacy and potential to tackle previously undrugged targets. Stable and long-lived degrader-mediated ternary complexes drive fast and profound target degradation; however, the mechanisms by which they affect target ubiquitination remain elusive. Here, we show cryo-EM structures of the VHL Cullin 2 RING E3 ligase with the degrader MZ1 directing target protein Brd4 toward UBE2R1-ubiquitin, and Lys at optimal positioning for nucleophilic attack. In vitro ubiquitination and mass spectrometry illuminate a patch of favorably ubiquitinable lysines on one face of Brd4, with cellular degradation and ubiquitinomics confirming the importance of Lys and nearby Lys/Lys, identifying the "ubiquitination zone." Our results demonstrate the proficiency of MZ1 in positioning the substrate for catalysis, the favorability of Brd4 for ubiquitination by UBE2R1, and the flexibility of CRL2 for capturing suboptimal lysines. We propose a model for ubiquitinability of degrader-recruited targets, providing a mechanistic blueprint for further rational drug design. #1:  Journal: Biorxiv / Year: 2024 Journal: Biorxiv / Year: 2024Title: Mechanism of degrader-targeted protein ubiquitinability Authors: Crowe C / Nakasone MA / Chandler S / Tatham MH / Makukhin N / Hay RT / Ciulli A | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_19567.map.gz emd_19567.map.gz | 398.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-19567-v30.xml emd-19567-v30.xml emd-19567.xml emd-19567.xml | 14.2 KB 14.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_19567_fsc.xml emd_19567_fsc.xml | 15.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_19567.png emd_19567.png | 110.9 KB | ||
| Filedesc metadata |  emd-19567.cif.gz emd-19567.cif.gz | 4.3 KB | ||
| Others |  emd_19567_half_map_1.map.gz emd_19567_half_map_1.map.gz emd_19567_half_map_2.map.gz emd_19567_half_map_2.map.gz | 391.3 MB 391.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-19567 http://ftp.pdbj.org/pub/emdb/structures/EMD-19567 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-19567 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-19567 | HTTPS FTP |
-Related structure data
| Related structure data |  8rx0MC  8rwzC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_19567.map.gz / Format: CCP4 / Size: 421.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_19567.map.gz / Format: CCP4 / Size: 421.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM structure of the (NEDD8)-CRL2VHL-MZ1-Brd4BD2 ternary complex primed for catalysis with UBE2R1-Ub. | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.825 Å | ||||||||||||||||||||||||||||||||||||
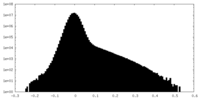
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_19567_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
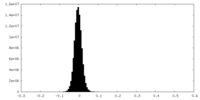
| Density Histograms |
-Half map: #2
| File | emd_19567_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
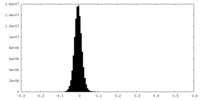
| Density Histograms |
- Sample components
Sample components
-Entire : Closed crosslinked structure of (NEDD8)-CRL2VHL-MZ1-Brd4BD2-Ub(G7...
| Entire | Name: Closed crosslinked structure of (NEDD8)-CRL2VHL-MZ1-Brd4BD2-Ub(G76S, K48C)-UBE2R1(C93K, S138C, C191S, C223S)-Ub |
|---|---|
| Components |
|
-Supramolecule #1: Closed crosslinked structure of (NEDD8)-CRL2VHL-MZ1-Brd4BD2-Ub(G7...
| Supramolecule | Name: Closed crosslinked structure of (NEDD8)-CRL2VHL-MZ1-Brd4BD2-Ub(G76S, K48C)-UBE2R1(C93K, S138C, C191S, C223S)-Ub type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 190 kDa/nm |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 38.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.7 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)