+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
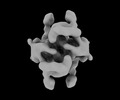
| Title | Cryo-EM structure of the DnaD-NTD tetramer | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Replication helicase loading / small cryo-EM structure / DNA BINDING PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology information | |||||||||
| Biological species |   | |||||||||
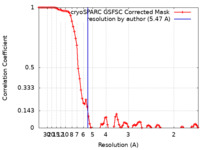
| Method | single particle reconstruction / cryo EM / Resolution: 5.47 Å | |||||||||
 Authors Authors | Winterhalter C / Pelliciari S / Cronin N / Costa TRD / Murray H / Ilangovan A | |||||||||
| Funding support |  United Kingdom, 1 items United Kingdom, 1 items
| |||||||||
 Citation Citation |  Journal: Nucleic Acids Res / Year: 2023 Journal: Nucleic Acids Res / Year: 2023Title: The DNA replication initiation protein DnaD recognises a specific strand of the Bacillus subtilis chromosome origin. Authors: Charles Winterhalter / Simone Pelliciari / Daniel Stevens / Stepan Fenyk / Elie Marchand / Nora B Cronin / Panos Soultanas / Tiago R D Costa / Aravindan Ilangovan / Heath Murray /  Abstract: Genome replication is a fundamental biological activity shared by all organisms. Chromosomal replication proceeds bidirectionally from origins, requiring the loading of two helicases, one for each ...Genome replication is a fundamental biological activity shared by all organisms. Chromosomal replication proceeds bidirectionally from origins, requiring the loading of two helicases, one for each replisome. However, the molecular mechanisms underpinning helicase loading at bacterial chromosome origins (oriC) are unclear. Here we investigated the essential DNA replication initiation protein DnaD in the model organism Bacillus subtilis. A set of DnaD residues required for ssDNA binding was identified, and photo-crosslinking revealed that this ssDNA binding region interacts preferentially with one strand of oriC. Biochemical and genetic data support the model that DnaD recognizes a new single-stranded DNA (ssDNA) motif located in oriC, the DnaD Recognition Element (DRE). Considered with single particle cryo-electron microscopy (cryo-EM) imaging of DnaD, we propose that the location of the DRE within oriC orchestrates strand-specific recruitment of helicase during DNA replication initiation. These findings significantly advance our mechanistic understanding of bidirectional replication from a bacterial chromosome origin. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_16914.map.gz emd_16914.map.gz | 59.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-16914-v30.xml emd-16914-v30.xml emd-16914.xml emd-16914.xml | 13.7 KB 13.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_16914_fsc.xml emd_16914_fsc.xml | 8.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_16914.png emd_16914.png | 33.7 KB | ||
| Filedesc metadata |  emd-16914.cif.gz emd-16914.cif.gz | 5.2 KB | ||
| Others |  emd_16914_half_map_1.map.gz emd_16914_half_map_1.map.gz emd_16914_half_map_2.map.gz emd_16914_half_map_2.map.gz | 59.5 MB 59.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-16914 http://ftp.pdbj.org/pub/emdb/structures/EMD-16914 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16914 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16914 | HTTPS FTP |
-Validation report
| Summary document |  emd_16914_validation.pdf.gz emd_16914_validation.pdf.gz | 727 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_16914_full_validation.pdf.gz emd_16914_full_validation.pdf.gz | 726.6 KB | Display | |
| Data in XML |  emd_16914_validation.xml.gz emd_16914_validation.xml.gz | 16.3 KB | Display | |
| Data in CIF |  emd_16914_validation.cif.gz emd_16914_validation.cif.gz | 21 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16914 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16914 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16914 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16914 | HTTPS FTP |
-Related structure data
| Related structure data |  8ojjMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_16914.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_16914.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.831 Å | ||||||||||||||||||||||||||||||||||||
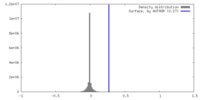
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_16914_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
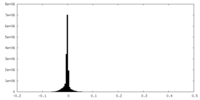
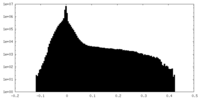
| Density Histograms |
-Half map: #2
| File | emd_16914_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
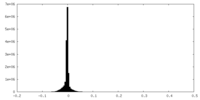
| Density Histograms |
- Sample components
Sample components
-Entire : Homo Tetrameric complex of DnaD N-terminal domain
| Entire | Name: Homo Tetrameric complex of DnaD N-terminal domain |
|---|---|
| Components |
|
-Supramolecule #1: Homo Tetrameric complex of DnaD N-terminal domain
| Supramolecule | Name: Homo Tetrameric complex of DnaD N-terminal domain / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: DNA replication protein DnaD
| Macromolecule | Name: DNA replication protein DnaD / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Strain: 168 |
| Molecular weight | Theoretical: 27.675715 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MKKQQFIDMQ EQGTSTIPNL LLTHYKQLGL NETELILLLK IKMHLEKGSY FPTPNQLQEG MSISVEECTN RLRMFIQKGF LFIEECEDQ NGIKFEKYSL QPLWGKLYEY IQLAQNQTQE RKAEGEQKSL YTIFEEEFAR PLSPLECETL AIWQDQDQHD A QLIKHALK ...String: MKKQQFIDMQ EQGTSTIPNL LLTHYKQLGL NETELILLLK IKMHLEKGSY FPTPNQLQEG MSISVEECTN RLRMFIQKGF LFIEECEDQ NGIKFEKYSL QPLWGKLYEY IQLAQNQTQE RKAEGEQKSL YTIFEEEFAR PLSPLECETL AIWQDQDQHD A QLIKHALK EAVLSGKLSF RYIDRILFEW KKNGLKTVEQ AKIHSQKFRR VQAKQNEPQK EYKRQVPFYN WLEQ UniProtKB: Replicative helicase loading/DNA remodeling protein DnaD |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.7 µm / Nominal defocus min: 0.7000000000000001 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Protocol: RIGID BODY FIT |
|---|---|
| Output model |  PDB-8ojj: |
 Movie
Movie Controller
Controller



 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)