3MHD
 
 | | Crystal structure of DCR3 | | Descriptor: | Tumor necrosis factor receptor superfamily member 6B | | Authors: | Zhan, C, Patskovsky, Y, Yan, Q, Li, Z, Ramagopal, U.A, Nathenson, S.G, Almo, S.C. | | Deposit date: | 2010-04-07 | | Release date: | 2011-02-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.901 Å) | | Cite: | Decoy Strategies: The Structure of TL1A:DcR3 Complex.
Structure, 19, 2011
|
|
3MI8
 
 | | The structure of TL1A-DCR3 COMPLEX | | Descriptor: | TUMOR NECROSIS FACTOR LIGAND SUPERFAMILY MEMBER 15, SECRETED FORM, Tumor necrosis factor receptor superfamily member 6B | | Authors: | Zhan, C, Patskovsky, Y, Yan, Q, Li, Z, Ramagopal, U.A, Nathenson, S.G, Almo, S.C. | | Deposit date: | 2010-04-09 | | Release date: | 2011-02-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.951 Å) | | Cite: | Decoy Strategies: The Structure of TL1A:DcR3 Complex.
Structure, 19, 2011
|
|
2QE3
 
 | | Crystal structure of human tl1a extracellular domain | | Descriptor: | CHLORIDE ION, TNF superfamily ligand TL1A | | Authors: | Zhan, C, Yan, Q, Patskovsky, Y, Shi, W, Toro, R, Bonanno, J, Nathenson, S.G, Almo, S.C. | | Deposit date: | 2007-06-22 | | Release date: | 2007-07-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Biochemical and structural characterization of the human TL1A ectodomain.
Biochemistry, 48, 2009
|
|
2RJL
 
 | | Crystal structure of human TL1A extracellular domain C95S/C135S mutant | | Descriptor: | TNF superfamily ligand TL1A | | Authors: | Zhan, C, Patskovsky, Y, Yan, Q, Shi, W, Toro, R, Bonanno, J, Nathenson, S.G, Almo, S.C. | | Deposit date: | 2007-10-15 | | Release date: | 2008-08-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Biochemical and structural characterization of the human TL1A ectodomain.
Biochemistry, 48, 2009
|
|
2RJK
 
 | | Crystal Structure of Human TL1A Extracellular Domain C95S Mutant | | Descriptor: | TNF superfamily ligand TL1A | | Authors: | Zhan, C, Yan, Q, Patskovsky, Y, Shi, W, Ramagopal, U.A, Toro, R, Bonanno, J, Nathenson, S.G, Almo, S.C. | | Deposit date: | 2007-10-15 | | Release date: | 2008-08-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Biochemical and structural characterization of the human TL1A ectodomain.
Biochemistry, 48, 2009
|
|
2HC9
 
 | | Structure of Caenorhabditis elegans leucine aminopeptidase-zinc complex (LAP1) | | Descriptor: | BICARBONATE ION, GLYCEROL, Leucine aminopeptidase 1, ... | | Authors: | Zhan, C, Patskovsky, Y, Wengerter, B.C, Ramagopal, U, Milstein, S, Vidal, M, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-06-15 | | Release date: | 2006-08-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure and Function of Caenorhabditis Elegans Leucine Aminopeptidase
To be Published
|
|
3K51
 
 | | Crystal Structure of DcR3-TL1A complex | | Descriptor: | Decoy receptor 3, Tumor necrosis factor ligand superfamily member 15, secreted form | | Authors: | Zhan, C, Patskovsky, Y, Yan, Q, Li, Z, Ramagopal, U.A, Nathenson, S.G, Almo, S.C. | | Deposit date: | 2009-10-06 | | Release date: | 2010-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Decoy Strategies: The Structure of TL1A:DcR3 Complex.
Structure, 19, 2011
|
|
8EPG
 
 | | Engineering Crystals with Tunable Symmetries from 14- or 16-Base-Long DNA Strands | | Descriptor: | DNA (5'-D(*CP*GP*AP*CP*G)-3'), DNA (5'-D(P*CP*GP*TP*GP*GP*C)-3'), DNA (5'-D(P*GP*CP*CP*GP*C)-3') | | Authors: | Zhang, C, Zhao, J, Lu, B, Sha, R, Seeman, N.C, Noinaj, N, Mao, C. | | Deposit date: | 2022-10-05 | | Release date: | 2023-03-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Engineering DNA Crystals toward Studying DNA-Guest Molecule Interactions.
J.Am.Chem.Soc., 145, 2023
|
|
8EPE
 
 | | Engineering Crystals with Tunable Symmetries from 14- or 16-Base-Long DNA Strands | | Descriptor: | DNA (5'-D(*CP*GP*AP*CP*G)-3'), DNA (5'-D(P*CP*CP*CP*GP*C)-3'), DNA (5'-D(P*CP*GP*TP*GP*GP*G)-3') | | Authors: | Zhang, C, Zhao, J, Lu, B, Sha, R, Seeman, N.C, Noinaj, N, Mao, C. | | Deposit date: | 2022-10-05 | | Release date: | 2023-03-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Engineering DNA Crystals toward Studying DNA-Guest Molecule Interactions.
J.Am.Chem.Soc., 145, 2023
|
|
8F42
 
 | | Engineering Crystals with Tunable Symmetries from 14- or 16-Base-Long DNA Strands | | Descriptor: | 2'-(4-ETHOXYPHENYL)-5-(4-METHYL-1-PIPERAZINYL)-2,5'-BI-BENZIMIDAZOLE, DNA (5'-D(*CP*GP*CP*TP*TP*AP*AP*GP*GP*AP*AP*TP*TP*CP*GP*C)-3') | | Authors: | Zhang, C, Zhao, J, Lu, B, Sha, R, Seeman, N.C, Noinaj, N, Mao, C. | | Deposit date: | 2022-11-10 | | Release date: | 2023-03-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Engineering DNA Crystals toward Studying DNA-Guest Molecule Interactions.
J.Am.Chem.Soc., 145, 2023
|
|
8EPB
 
 | | Engineering Crystals with Tunable Symmetries from 14- or 16-Base-Long DNA Strands | | Descriptor: | DNA (5'-D(*CP*GP*CP*TP*GP*GP*TP*GP*GP*TP*TP*CP*GP*A)-3'), DNA (5'-D(*GP*AP*CP*CP*AP*GP*CP*CP*GP*AP*AP*CP*CP*T)-3') | | Authors: | Zhang, C, Zhao, J, Lu, B, Sha, R, Seeman, N.C, Noinaj, N, Mao, C. | | Deposit date: | 2022-10-05 | | Release date: | 2023-03-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Engineering DNA Crystals toward Studying DNA-Guest Molecule Interactions.
J.Am.Chem.Soc., 145, 2023
|
|
8EPI
 
 | | Engineering Crystals with Tunable Symmetries from 14- or 16-Base-Long DNA Strands | | Descriptor: | DNA (5'-D(*CP*GP*AP*CP*G)-3'), DNA (5'-D(P*CP*GP*TP*GP*GP*A)-3'), DNA (5'-D(P*TP*CP*CP*TP*A)-3') | | Authors: | Zhang, C, Zhao, J, Lu, B, Sha, R, Seeman, N.C, Noinaj, N, Mao, C. | | Deposit date: | 2022-10-05 | | Release date: | 2023-03-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Engineering DNA Crystals toward Studying DNA-Guest Molecule Interactions.
J.Am.Chem.Soc., 145, 2023
|
|
8F40
 
 | | Engineering Crystals with Tunable Symmetries from 14- or 16-Base-Long DNA Strands | | Descriptor: | DNA (5'-D(*CP*GP*CP*TP*T)-3'), DNA (5'-D(P*AP*AP*GP*GP*AP*A)-3'), DNA (5'-D(P*TP*TP*CP*GP*C)-3') | | Authors: | Zhang, C, Zhao, J, Lu, B, Sha, R, Seeman, N.C, Noinaj, N, Mao, C. | | Deposit date: | 2022-11-10 | | Release date: | 2023-03-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Engineering DNA Crystals toward Studying DNA-Guest Molecule Interactions.
J.Am.Chem.Soc., 145, 2023
|
|
8EPD
 
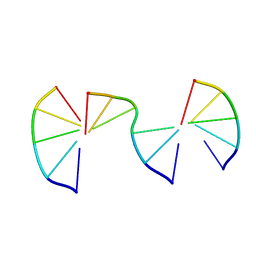 | | Engineering Crystals with Tunable Symmetries from 14- or 16-Base-Long DNA Strands | | Descriptor: | DNA (5'-D(*CP*GP*AP*CP*G)-3'), DNA (5'-D(P*CP*GP*TP*TP*CP*C)-3'), DNA (5'-D(P*GP*GP*AP*GP*C)-3') | | Authors: | Zhang, C, Zhao, J, Lu, B, Sha, R, Seeman, N.C, Noinaj, N, Mao, C. | | Deposit date: | 2022-10-05 | | Release date: | 2023-03-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Engineering DNA Crystals toward Studying DNA-Guest Molecule Interactions.
J.Am.Chem.Soc., 145, 2023
|
|
8EPF
 
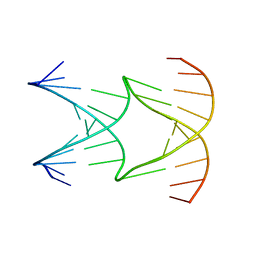 | | Engineering Crystals with Tunable Symmetries from 14- or 16-Base-Long DNA Strands | | Descriptor: | DNA (5'-D(*AP*CP*GP*CP*TP*GP*GP*TP*GP*GP*TP*TP*CP*GP*CP*A)-3'), DNA (5'-D(*GP*TP*AP*CP*CP*AP*GP*CP*CP*GP*AP*AP*CP*CP*TP*G)-3') | | Authors: | Zhang, C, Zhao, J, Lu, B, Sha, R, Seeman, N.C, Noinaj, N, Mao, C. | | Deposit date: | 2022-10-05 | | Release date: | 2023-03-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Engineering DNA Crystals toward Studying DNA-Guest Molecule Interactions.
J.Am.Chem.Soc., 145, 2023
|
|
8EP8
 
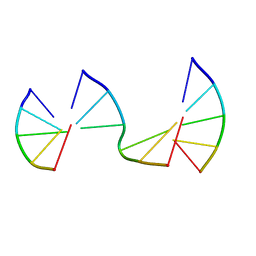 | | Engineering Crystals with Tunable Symmetries from 14- or 16-Base-Long DNA Strands | | Descriptor: | DNA (5'-D(*CP*GP*AP*CP*G)-3'), DNA (5'-D(P*CP*GP*TP*GP*GP*A)-3'), DNA (5'-D(P*TP*CP*CP*GP*C)-3') | | Authors: | Zhang, C, Zhao, J, Lu, B, Sha, R, Seeman, N.C, Noinaj, N, Mao, C. | | Deposit date: | 2022-10-05 | | Release date: | 2023-03-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Engineering DNA Crystals toward Studying DNA-Guest Molecule Interactions.
J.Am.Chem.Soc., 145, 2023
|
|
8GH5
 
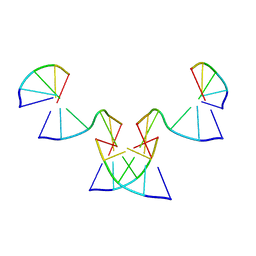 | | Implementing Logic Gates in DNA Crystal Engineering | | Descriptor: | DNA (5'-D(*AP*GP*AP*CP*G)-3'), DNA (5'-D(*CP*TP*AP*CP*G)-3'), DNA (5'-D(P*CP*GP*TP*GP*GP*A)-3'), ... | | Authors: | Zhang, C, Paluzzi, V.E, Sha, R, Jonoska, N, Mao, C. | | Deposit date: | 2023-03-09 | | Release date: | 2023-06-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Implementing Logic Gates by DNA Crystal Engineering.
Adv Mater, 35, 2023
|
|
7RIK
 
 | | Magic-Angle-Spinning NMR Structure of Kinesin-1 Motor Domain Assembled with Microtubules | | Descriptor: | Kinesin-1 heavy chain | | Authors: | Zhang, C, Guo, C, Russell, R.W, Quinn, C.M, Li, M, Williams, J.C, Gronenborn, A.M, Polenova, T. | | Deposit date: | 2021-07-20 | | Release date: | 2022-11-23 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Magic-angle-spinning NMR structure of the kinesin-1 motor domain assembled with microtubules reveals the elusive neck linker orientation
Nat Commun, 13, 2022
|
|
2DHO
 
 | | Crystal structure of human IPP isomerase I in space group P212121 | | Descriptor: | CHLORIDE ION, Isopentenyl-diphosphate delta-isomerase 1, MANGANESE (II) ION, ... | | Authors: | Zhang, C, Wei, Z, Gong, W. | | Deposit date: | 2006-03-24 | | Release date: | 2007-06-05 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of human IPP isomerase: new insights into the catalytic mechanism
J.Mol.Biol., 366, 2007
|
|
5ICA
 
 | | Structure of the CTD complex of UTP12, Utp13, Utp1 and Utp21 | | Descriptor: | Periodic tryptophan protein 2-like protein, Putative U3 snoRNP protein, Putative uncharacterized protein | | Authors: | Zhang, C, Ye, K. | | Deposit date: | 2016-02-23 | | Release date: | 2016-07-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.507 Å) | | Cite: | Integrative structural analysis of the UTPB complex, an early assembly factor for eukaryotic small ribosomal subunits
Nucleic Acids Res., 44, 2016
|
|
5GP1
 
 | | Crystal structure of ZIKV NS5 Methyltransferase in complex with GTP and SAH | | Descriptor: | NICKEL (II) ION, P1-7-METHYLGUANOSINE-P3-ADENOSINE-5',5'-TRIPHOSPHATE, RNA-directed RNA polymerase NS5, ... | | Authors: | Zhang, C, Jin, T. | | Deposit date: | 2016-07-30 | | Release date: | 2016-12-07 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.444 Å) | | Cite: | Structure of the NS5 methyltransferase from Zika virus and implications in inhibitor design
Biochem. Biophys. Res. Commun., 492, 2017
|
|
5GOZ
 
 | |
5IC7
 
 | | Structure of the WD domain of UTP18 | | Descriptor: | Putative uncharacterized protein, SULFATE ION | | Authors: | Zhang, C, Ye, K. | | Deposit date: | 2016-02-22 | | Release date: | 2016-07-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.331 Å) | | Cite: | Integrative structural analysis of the UTPB complex, an early assembly factor for eukaryotic small ribosomal subunits
Nucleic Acids Res., 44, 2016
|
|
5IC9
 
 | | Structure of the CTD complex of Utp12 and Utp13 | | Descriptor: | Putative uncharacterized protein | | Authors: | Zhang, C, Ye, K. | | Deposit date: | 2016-02-23 | | Release date: | 2016-07-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Integrative structural analysis of the UTPB complex, an early assembly factor for eukaryotic small ribosomal subunits
Nucleic Acids Res., 44, 2016
|
|
5IC8
 
 | | Structure of UTP6 | | Descriptor: | Putative uncharacterized protein | | Authors: | Zhang, C, Ye, K. | | Deposit date: | 2016-02-23 | | Release date: | 2016-07-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Integrative structural analysis of the UTPB complex, an early assembly factor for eukaryotic small ribosomal subunits
Nucleic Acids Res., 44, 2016
|
|
