1E54
 
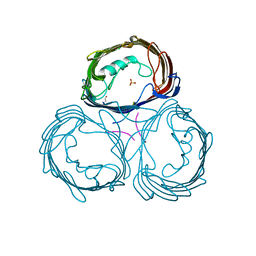 | | Anion-selective porin from Comamonas acidovorans | | Descriptor: | CALCIUM ION, OMP32, OUTER MEMBRANE PORIN PROTEIN 32, ... | | Authors: | Zeth, K, Diederichs, K, Welte, W, Engelhardt, H. | | Deposit date: | 2000-07-17 | | Release date: | 2001-07-12 | | Last modified: | 2020-03-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Omp32, the Anion-Selective Porin from Comamonas Acidovorans, in Complex with a Periplasmic Peptideat 2.1 A Resolution
Structure, 8, 2000
|
|
6SEV
 
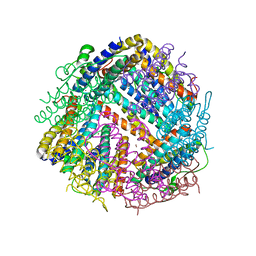 | |
1LZW
 
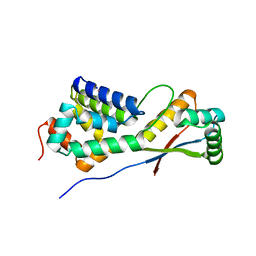 | | Structural basis of ClpS-mediated switch in ClpA substrate recognition | | Descriptor: | ATP-dependent clp protease ATP-binding subunit ClpA, PLATINUM (II) ION, Protein yljA | | Authors: | Zeth, K, Ravelli, R.B, Paal, K, Cusack, S, Bukau, B, Dougan, D.A. | | Deposit date: | 2002-06-11 | | Release date: | 2002-11-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural analysis of the adaptor protein ClpS in complex with the N-terminal domain of ClpA
Nat.Struct.Biol., 9, 2002
|
|
1MG9
 
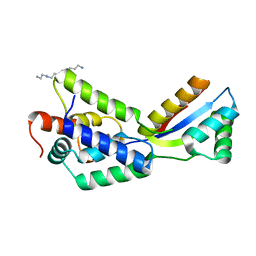 | | The structural basis of ClpS-mediated switch in ClpA substrate recognition | | Descriptor: | ATP dependent clp protease ATP-binding subunit clpA, SPERMINE (FULLY PROTONATED FORM), protein yljA | | Authors: | Zeth, K, Ravelli, R.B, Paal, K, Cusack, S, Bukau, B, Dougan, D.A. | | Deposit date: | 2002-08-15 | | Release date: | 2002-11-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural analysis of the adaptor protein ClpS in complex with the N-terminal domain of ClpA
Nat.Struct.Biol., 9, 2002
|
|
1MOJ
 
 | | Crystal structure of an archaeal dps-homologue from Halobacterium salinarum | | Descriptor: | Dps-like ferritin, FE (III) ION, MAGNESIUM ION, ... | | Authors: | Zeth, K, Offermann, S, Essen, L.O, Oesterhelt, D. | | Deposit date: | 2002-09-09 | | Release date: | 2004-04-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Iron-oxo clusters biomineralizing on protein surfaces: Structural analysis of Halobacterium salinarum DpsA in its low- and high-iron states.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
6HX2
 
 | |
6HUI
 
 | |
6HV1
 
 | |
6HVQ
 
 | |
3DA3
 
 | | Crystal Structure of Colicin M, A Novel Phosphatase Specifically Imported by Escherichia Coli | | Descriptor: | Colicin-M, MAGNESIUM ION | | Authors: | Zeth, K, Albrecht, R, Romer, C, Braun, V. | | Deposit date: | 2008-05-28 | | Release date: | 2008-09-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of colicin M, a novel phosphatase specifically imported by Escherichia coli
J.Biol.Chem., 283, 2008
|
|
3DA4
 
 | | Crystal Structure of Colicin M, a Novel Phosphatase Specifically Imported by Escherichia Coli | | Descriptor: | Colicin-M, NITRATE ION | | Authors: | Zeth, K, Albrecht, R, Romer, C, Braun, V. | | Deposit date: | 2008-05-28 | | Release date: | 2008-07-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of colicin M, a novel phosphatase specifically imported by Escherichia coli
J.Biol.Chem., 283, 2008
|
|
1TJO
 
 | | Iron-oxo clusters biomineralizing on protein surfaces. Structural analysis of H.salinarum DpsA in its low and high iron states | | Descriptor: | FE (III) ION, Iron-rich dpsA-homolog protein, MAGNESIUM ION, ... | | Authors: | Zeth, K, Offermann, S, Essen, L.O, Oesterhelt, D. | | Deposit date: | 2004-06-07 | | Release date: | 2004-10-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Iron-oxo clusters biomineralizing on protein surfaces: structural analysis of Halobacterium salinarum DpsA in its low- and high-iron states.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1TK6
 
 | | Iron-oxo clusters biomineralizing on protein surfaces. Structural analysis of H.salinarum DpsA in its low and high iron states | | Descriptor: | FE (III) ION, Iron-rich dpsA-homolog protein, MAGNESIUM ION, ... | | Authors: | Zeth, K, Offermann, S, Essen, L.O, Oesterhelt, D. | | Deposit date: | 2004-06-08 | | Release date: | 2004-10-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Iron-oxo clusters biomineralizing on protein surfaces: structural analysis of Halobacterium salinarum DpsA in its low- and high-iron states.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1TKO
 
 | | Iron-oxo clusters biomineralizing on protein surfaces. Structural analysis of H.salinarum DpsA in its low and high iron states | | Descriptor: | FE (III) ION, Iron-rich dpsA-homolog protein, SODIUM ION, ... | | Authors: | Zeth, K, Offermann, S, Essen, L.O, Oesterhelt, D. | | Deposit date: | 2004-06-09 | | Release date: | 2004-10-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Iron-oxo clusters biomineralizing on protein surfaces: structural analysis of Halobacterium salinarum DpsA in its low- and high-iron states.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1TKP
 
 | | Iron-oxo clusters biomineralizing on protein surfaces. Structural analysis of H.salinarum DpsA in its low and high iron states | | Descriptor: | FE (III) ION, Iron-rich dpsA-homolog protein, SODIUM ION, ... | | Authors: | Zeth, K, Offermann, S, Essen, L.O, Oesterhelt, D. | | Deposit date: | 2004-06-09 | | Release date: | 2004-10-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Iron-oxo clusters biomineralizing on protein surfaces: structural analysis of Halobacterium salinarum DpsA in its low- and high-iron states.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
4AFF
 
 | | High resolution structure of a PII mutant (I86N) protein in complex with ATP, MG and FLC | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CITRATE ANION, MAGNESIUM ION, ... | | Authors: | Zeth, K, Fokina, O, Chellamuthu, V.R, Forchhammer, K. | | Deposit date: | 2012-01-19 | | Release date: | 2012-02-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | An Engineered Pii Protein Variant that Senses a Novel Ligand: Atomic Resolution Structure of the Complex with Citrate.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4C3L
 
 | | Structure of wildtype PII from S. elongatus at high resolution | | Descriptor: | CHLORIDE ION, NITROGEN REGULATORY PROTEIN P-II, TETRAETHYLENE GLYCOL | | Authors: | Zeth, K, Forchhammer, K. | | Deposit date: | 2013-08-25 | | Release date: | 2014-01-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Basis and Target-Specific Modulation of Adp Sensing by the Synechococcus Elongatus Pii Signaling Protein.
J.Biol.Chem., 289, 2014
|
|
4C3K
 
 | | Structure of mixed PII-ADP complexes from S. elongatus | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, NITROGEN REGULATORY PROTEIN P-II, PHOSPHATE ION | | Authors: | Zeth, K, Forchhammer, K. | | Deposit date: | 2013-08-25 | | Release date: | 2014-02-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.099 Å) | | Cite: | Structural Basis and Target-Specific Modulation of Adp Sensing by the Synechococcus Elongatus Pii Signaling Protein.
J.Biol.Chem., 289, 2014
|
|
4C3M
 
 | |
4AUI
 
 | | STRUCTURE AND FUNCTION OF THE PORB PORIN FROM DISSEMINATING N. GONORRHOEAE | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Zeth, K, Kozjak-Pavlovic, V, Faulstich, M, Hurwitz, R, Kepp, O, Rudel, T. | | Deposit date: | 2012-05-17 | | Release date: | 2012-10-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure and Function of the Porb Porin from Disseminating Neisseria Gonorrhoeae.
Biochem.J., 449, 2013
|
|
3ZCC
 
 | |
4BAX
 
 | |
2YLE
 
 | | Crystal structure of the human Spir-1 KIND FSI domain in complex with the FSI peptide | | Descriptor: | FORMIN-2, PROTEIN SPIRE HOMOLOG 1 | | Authors: | Zeth, K, Pechlivanis, M, Vonrhein, C, Kerkhoff, E. | | Deposit date: | 2011-06-01 | | Release date: | 2011-06-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular Basis of Actin Nucleation Factor Cooperativity: Crystal Structure of the Spir-1 Kinase Non-Catalytic C-Lobe Domain (Kind)Formin-2 Formin Spir Interaction Motif (Fsi) Complex.
J.Biol.Chem., 286, 2011
|
|
2YLF
 
 | |
2XUL
 
 | | Structure of PII from Synechococcus elongatus in complex with 2- oxoglutarate at high 2-OG concentrations | | Descriptor: | 2-OXOGLUTARIC ACID, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Zeth, K, Chellamuthu, V.-R, Forchhammer, K, Fokina, O. | | Deposit date: | 2010-10-19 | | Release date: | 2010-11-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mechanism of 2-Oxoglutarate Signaling by the Synechococcus Elongatus Pii Signal Transduction Protein
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
