8QOT
 
 | | Structure of the mu opioid receptor bound to the antagonist nanobody NbE | | Descriptor: | Anti-Fab Nanobody, Mu-type opioid receptor, NabFab HC, ... | | Authors: | Yu, J, Kumar, A, Zhang, X, Martin, C, Raia, P, Manglik, A, Ballet, S, Boland, A, Stoeber, M. | | Deposit date: | 2023-09-29 | | Release date: | 2023-12-27 | | Last modified: | 2025-01-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis of mu-opioid receptor targeting by a nanobody antagonist.
Nat Commun, 15, 2024
|
|
7NJ0
 
 | | CryoEM structure of the human Separase-Cdk1-cyclin B1-Cks1 complex | | Descriptor: | Cyclin-dependent kinase 1, Cyclin-dependent kinases regulatory subunit 1, G2/mitotic-specific cyclin-B1,G2/mitotic-specific cyclin-B1, ... | | Authors: | Yu, J, Raia, P, Ghent, C.M, Raisch, T, Sadian, Y, Barford, D, Raunser, S, Morgan, D.O, Boland, A. | | Deposit date: | 2021-02-14 | | Release date: | 2021-08-04 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis of human separase regulation by securin and CDK1-cyclin B1.
Nature, 596, 2021
|
|
7NJ1
 
 | | CryoEM structure of the human Separase-Securin complex | | Descriptor: | Securin, Separin | | Authors: | Yu, J, Raia, P, Ghent, C.M, Raisch, T, Sadian, Y, Barford, D, Raunser, S, Morgan, D.O, Boland, A. | | Deposit date: | 2021-02-14 | | Release date: | 2021-08-04 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of human separase regulation by securin and CDK1-cyclin B1.
Nature, 596, 2021
|
|
4XZ7
 
 | | Crystal structure of a TGase | | Descriptor: | Putative uncharacterized protein | | Authors: | Yu, J, Ge, J, Yang, M. | | Deposit date: | 2015-02-04 | | Release date: | 2015-06-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Functional and Structural Characterization of the Antiphagocytic Properties of a Novel Transglutaminase from Streptococcus suis
J.Biol.Chem., 290, 2015
|
|
8X19
 
 | | Structure of nucleosome-bound SRCAP-C in the ADP-BeFx-bound state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Actin, ... | | Authors: | Yu, J, Wang, Q, Yu, Z, Li, W, Wang, L, Xu, Y. | | Deposit date: | 2023-11-06 | | Release date: | 2024-03-06 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into histone exchange by human SRCAP complex.
Cell Discov, 10, 2024
|
|
8X15
 
 | | Structure of nucleosome-bound SRCAP-C in the apo state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Actin, ... | | Authors: | Yu, J, Wang, Q, Yu, Z, Li, W, Wang, L, Xu, Y. | | Deposit date: | 2023-11-06 | | Release date: | 2024-03-06 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into histone exchange by human SRCAP complex.
Cell Discov, 10, 2024
|
|
8X1C
 
 | | Structure of nucleosome-bound SRCAP-C in the ADP-bound state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Actin, ... | | Authors: | Yu, J, Wang, Q, Yu, Z, Li, W, Wang, L, Xu, Y. | | Deposit date: | 2023-11-06 | | Release date: | 2024-03-06 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into histone exchange by human SRCAP complex.
Cell Discov, 10, 2024
|
|
6PLX
 
 | | CryoEM structure of zebra fish alpha-1 glycine receptor bound with GABA in SMA, desensitized state | | Descriptor: | GAMMA-AMINO-BUTANOIC ACID, Glycine receptor subunit alphaZ1, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Yu, J, Zhu, H, Gouaux, E. | | Deposit date: | 2019-07-01 | | Release date: | 2021-01-06 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Mechanism of gating and partial agonist action in the glycine receptor.
Cell, 184, 2021
|
|
6PLZ
 
 | | CryoEM structure of zebra fish alpha-1 glycine receptor bound with GABA in SMA, closed state | | Descriptor: | GAMMA-AMINO-BUTANOIC ACID, Glycine receptor subunit alphaZ1, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Yu, J, Zhu, H, Gouaux, E. | | Deposit date: | 2019-07-01 | | Release date: | 2021-01-06 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Mechanism of gating and partial agonist action in the glycine receptor.
Cell, 184, 2021
|
|
6PLY
 
 | | CryoEM structure of zebra fish alpha-1 glycine receptor bound with GABA in SMA, open state | | Descriptor: | GAMMA-AMINO-BUTANOIC ACID, Glycine receptor subunit alphaZ1, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Yu, J, Zhu, H, Gouaux, E. | | Deposit date: | 2019-07-01 | | Release date: | 2021-01-06 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Mechanism of gating and partial agonist action in the glycine receptor.
Cell, 184, 2021
|
|
6PLR
 
 | | CryoEM structure of zebra fish alpha-1 glycine receptor bound with glycine in nanodisc, desensitized state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCINE, Glycine receptor subunit alphaZ1, ... | | Authors: | Yu, J, Zhu, H, Gouaux, E. | | Deposit date: | 2019-07-01 | | Release date: | 2021-01-06 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mechanism of gating and partial agonist action in the glycine receptor.
Cell, 184, 2021
|
|
6PLO
 
 | | CryoEM structure of zebra fish alpha-1 glycine receptor YGF mutant bound with GABA in SMA,open state | | Descriptor: | GAMMA-AMINO-BUTANOIC ACID, Glycine receptor subunit alphaZ1, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Yu, J, Zhu, H, Gouaux, E. | | Deposit date: | 2019-07-01 | | Release date: | 2021-01-06 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Mechanism of gating and partial agonist action in the glycine receptor.
Cell, 184, 2021
|
|
6PLT
 
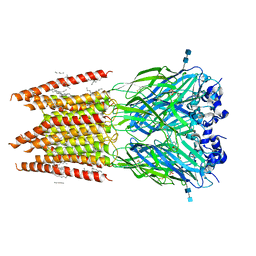 | | CryoEM structure of zebra fish alpha-1 glycine receptor bound with taurine in nanodisc, closed state | | Descriptor: | 2-AMINOETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glycine receptor subunit alphaZ1, ... | | Authors: | Yu, J, Zhu, H, Gouaux, E. | | Deposit date: | 2019-07-01 | | Release date: | 2021-01-06 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mechanism of gating and partial agonist action in the glycine receptor.
Cell, 184, 2021
|
|
6PLS
 
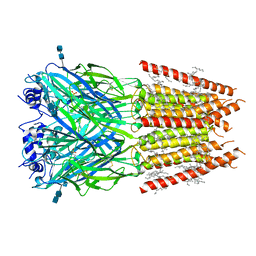 | | CryoEM structure of zebra fish alpha-1 glycine receptor bound with taurine in nanodisc, desensitized state | | Descriptor: | 2-AMINOETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glycine receptor subunit alphaZ1, ... | | Authors: | Yu, J, Zhu, H, Gouaux, E. | | Deposit date: | 2019-07-01 | | Release date: | 2021-01-06 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Mechanism of gating and partial agonist action in the glycine receptor.
Cell, 184, 2021
|
|
6PLV
 
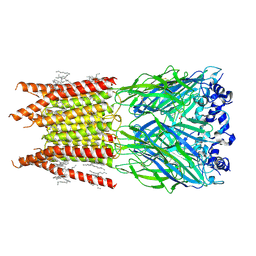 | | CryoEM structure of zebra fish alpha-1 glycine receptor bound with GABA in nanodisc, closed state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GAMMA-AMINO-BUTANOIC ACID, Glycine receptor subunit alphaZ1, ... | | Authors: | Yu, J, Zhu, H, Gouaux, E. | | Deposit date: | 2019-07-01 | | Release date: | 2021-01-06 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Mechanism of gating and partial agonist action in the glycine receptor.
Cell, 184, 2021
|
|
6PLU
 
 | | CryoEM structure of zebra fish alpha-1 glycine receptor bound with GABA in nanodisc, desensitized state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GAMMA-AMINO-BUTANOIC ACID, Glycine receptor subunit alphaZ1, ... | | Authors: | Yu, J, Zhu, H, Gouaux, E. | | Deposit date: | 2019-07-01 | | Release date: | 2021-01-06 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Mechanism of gating and partial agonist action in the glycine receptor.
Cell, 184, 2021
|
|
6PLQ
 
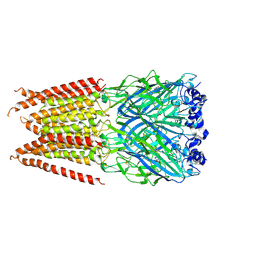 | | CryoEM structure of zebra fish alpha-1 glycine receptor YGF mutant bound with GABA in SMA, super-open state | | Descriptor: | GAMMA-AMINO-BUTANOIC ACID, Glycine receptor subunit alphaZ1, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Yu, J, Zhu, H, Gouaux, E. | | Deposit date: | 2019-07-01 | | Release date: | 2021-02-10 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Mechanism of gating and partial agonist action in the glycine receptor.
Cell, 184, 2021
|
|
6PM1
 
 | | CryoEM structure of zebra fish alpha-1 glycine receptor bound with Taurine in SMA, desensitized state | | Descriptor: | 2-AMINOETHANESULFONIC ACID, Glycine receptor subunit alphaZ1, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Yu, J, Zhu, H, Gouaux, E. | | Deposit date: | 2019-07-01 | | Release date: | 2021-02-10 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Mechanism of gating and partial agonist action in the glycine receptor.
Cell, 184, 2021
|
|
6PM0
 
 | | CryoEM structure of zebra fish alpha-1 glycine receptor bound with Taurine in SMA, super-open state | | Descriptor: | 2-AMINOETHANESULFONIC ACID, Glycine receptor subunit alphaZ1, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Yu, J, Zhu, H, Gouaux, E. | | Deposit date: | 2019-07-01 | | Release date: | 2021-02-10 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Mechanism of gating and partial agonist action in the glycine receptor.
Cell, 184, 2021
|
|
6PM4
 
 | | CryoEM structure of zebra fish alpha-1 glycine receptor bound with Glycine in SMA, super-open state | | Descriptor: | GLYCINE, Glycine receptor subunit alphaZ1, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Yu, J, Zhu, H, Gouaux, E. | | Deposit date: | 2019-07-01 | | Release date: | 2021-02-10 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Mechanism of gating and partial agonist action in the glycine receptor.
Cell, 184, 2021
|
|
6PM2
 
 | | CryoEM structure of zebra fish alpha-1 glycine receptor bound with Taurine in SMA, open state | | Descriptor: | 2-AMINOETHANESULFONIC ACID, Glycine receptor subunit alphaZ1, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Yu, J, Zhu, H, Gouaux, E. | | Deposit date: | 2019-07-01 | | Release date: | 2021-02-10 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Mechanism of gating and partial agonist action in the glycine receptor.
Cell, 184, 2021
|
|
6PM3
 
 | | CryoEM structure of zebra fish alpha-1 glycine receptor bound with Taurine in SMA, closed state | | Descriptor: | 2-AMINOETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glycine receptor subunit alphaZ1 | | Authors: | Yu, J, Zhu, H, Gouaux, E. | | Deposit date: | 2019-07-01 | | Release date: | 2021-02-10 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Mechanism of gating and partial agonist action in the glycine receptor.
Cell, 184, 2021
|
|
6PM5
 
 | | CryoEM structure of zebra fish alpha-1 glycine receptor bound with Glycine in SMA, desensitized state | | Descriptor: | GLYCINE, Glycine receptor subunit alphaZ1, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Yu, J, Zhu, H, Gouaux, E. | | Deposit date: | 2019-07-01 | | Release date: | 2021-02-10 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Mechanism of gating and partial agonist action in the glycine receptor.
Cell, 184, 2021
|
|
6PLP
 
 | | CryoEM structure of zebra fish alpha-1 glycine receptor YGF mutant bound with GABA in SMA, desensitized state | | Descriptor: | GAMMA-AMINO-BUTANOIC ACID, Glycine receptor subunit alphaZ1, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Yu, J, Zhu, H, Gouaux, E. | | Deposit date: | 2019-07-01 | | Release date: | 2021-02-10 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Mechanism of gating and partial agonist action in the glycine receptor.
Cell, 184, 2021
|
|
6PM6
 
 | | CryoEM structure of zebra fish alpha-1 glycine receptor bound with Glycine in SMA, open state | | Descriptor: | GLYCINE, Glycine receptor subunit alphaZ1, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Yu, J, Zhu, H, Gouaux, E. | | Deposit date: | 2019-07-01 | | Release date: | 2021-02-10 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Mechanism of gating and partial agonist action in the glycine receptor.
Cell, 184, 2021
|
|
