1TY4
 
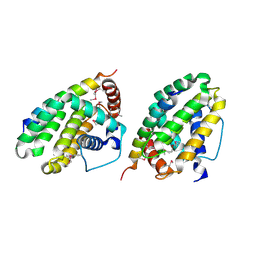 | | Crystal structure of a CED-9/EGL-1 complex | | Descriptor: | Apoptosis regulator ced-9, EGg Laying defective EGL-1, programmed cell death activator | | Authors: | Yan, N, Gu, L, Kokel, D, Xue, D, Shi, Y. | | Deposit date: | 2004-07-07 | | Release date: | 2004-09-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural, Biochemical, and Functional Analyses of CED-9 Recognition by the Proapoptotic Proteins EGL-1 and CED-4
Mol.Cell, 15, 2004
|
|
1SDZ
 
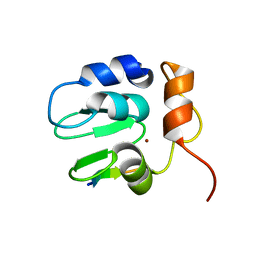 | | Crystal structure of DIAP1 BIR1 bound to a Reaper peptide | | Descriptor: | Apoptosis 1 inhibitor, Reaper, ZINC ION | | Authors: | Yan, N, Wu, J.W, Shi, Y. | | Deposit date: | 2004-02-15 | | Release date: | 2004-04-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Molecular mechanisms of DrICE inhibition by DIAP1 and removal of inhibition by Reaper, Hid and Grim.
Nat.Struct.Mol.Biol., 11, 2004
|
|
1SE0
 
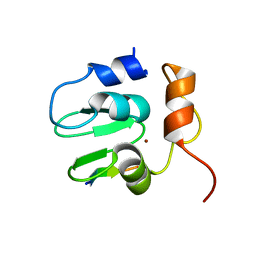 | | Crystal structure of DIAP1 BIR1 bound to a Grim peptide | | Descriptor: | Apoptosis 1 inhibitor, Cell death protein Grim, ZINC ION | | Authors: | Yan, N, Wu, J.W, Shi, Y. | | Deposit date: | 2004-02-15 | | Release date: | 2004-04-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Molecular mechanisms of DrICE inhibition by DIAP1 and removal of inhibition by Reaper, Hid and Grim.
Nat.Struct.Mol.Biol., 11, 2004
|
|
4GJP
 
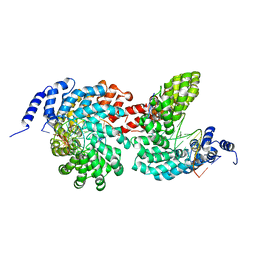 | | Crystal structure of the TAL effector dHax3 bound to dsDNA containing repetitive methyl-CpG | | Descriptor: | DNA (5'-D(*AP*GP*AP*GP*AP*CP*GP*CP*GP*AP*AP*GP*GP*GP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*CP*CP*CP*TP*TP*(5CM)P*GP*(5CM)P*GP*TP*CP*TP*CP*T)-3'), Hax3, ... | | Authors: | Yan, N, Deng, D, Yan, C.Y, Yin, P, Pan, X.J, Shi, Y.G. | | Deposit date: | 2012-08-10 | | Release date: | 2012-10-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal structure of a protein complex
To be Published
|
|
4GJR
 
 | | Crystal structure of the TAL effector dHax3 bound to methylated dsDNA | | Descriptor: | DNA (5'-D(*AP*GP*GP*GP*AP*GP*GP*TP*AP*GP*AP*GP*GP*GP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*CP*CP*CP*TP*(5CM)P*TP*AP*(5CM)P*CP*TP*CP*(5CM)P*CP*T)-3'), Hax3 | | Authors: | Yan, N, Deng, D, Yan, C.Y, Yin, P, Pan, X.J, Shi, Y.G. | | Deposit date: | 2012-08-10 | | Release date: | 2012-10-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of a protein complex
To be Published
|
|
2A5Y
 
 | | Structure of a CED-4/CED-9 complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Apoptosis regulator ced-9, MAGNESIUM ION, ... | | Authors: | Yan, N, Liu, Q, Hao, Q, Gu, L, Shi, Y. | | Deposit date: | 2005-07-01 | | Release date: | 2005-10-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the CED-4-CED-9 complex provides insights into programmed cell death in Caenorhabditis elegans.
Nature, 437, 2005
|
|
2FP3
 
 | |
4V2T
 
 | | Membrane embedded pleurotolysin pore with 13 fold symmetry | | Descriptor: | PLEUROTOLYSIN A, PLEUROTOLYSIN B | | Authors: | Lukoyanova, N, Kondos, S.C, Farabella, I, Law, R.H.P, Reboul, C.F, Caradoc-Davies, T.T, Spicer, B.A, Kleifeld, O, Perugini, M, Ekkel, S, Hatfaludi, T, Oliver, K, Hotze, E.M, Tweten, R.K, Whisstock, J.C, Topf, M, Dunstone, M.A, Saibil, H.R. | | Deposit date: | 2014-10-15 | | Release date: | 2015-02-18 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (11 Å) | | Cite: | Conformational Changes During Pore Formation by the Perforin-Related Protein Pleurotolysin.
Plos Biol., 13, 2015
|
|
4V3A
 
 | | Membrane bound pleurotolysin prepore (TMH1 lock) trapped with engineered disulphide cross-link | | Descriptor: | PLEUROTOLYSIN A, PLEUROTOLYSIN B | | Authors: | Lukoyanova, N, Kondos, S.C, Farabella, I, Law, R.H.P, Reboul, C.F, CaradocDavies, T.T, Spicer, B.A, Kleifeld, O, Perugini, M, Ekkel, S, Hatfaludi, T, Oliver, K, Hotze, E.M, Tweten, R.K, Whisstock, J.C, Topf, M, Dunstone, M.A, Saibil, H.R. | | Deposit date: | 2014-10-17 | | Release date: | 2015-02-18 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (15 Å) | | Cite: | Conformational Changes During Pore Formation by the Perforin-Related Protein Pleurotolysin.
Plos Biol., 13, 2015
|
|
4V3M
 
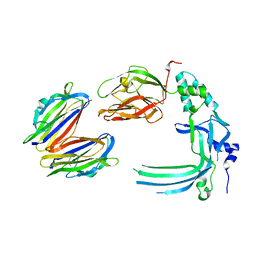 | | Membrane bound pleurotolysin prepore (TMH2 helix lock) trapped with engineered disulphide cross-link | | Descriptor: | PLEUROTOLYSIN A, PLEUROTOLYSIN B | | Authors: | Lukoyanova, N, Kondos, S.C, Farabella, I, Law, R.H.P, Reboul, C.F, Caradoc-Davies, T.T, Spicer, B.A, Kleifeld, O, Perugini, M, Ekkel, S, Hatfaludi, T, Oliver, K, Hotze, E.M, Tweten, R.K, Whisstock, J.C, Topf, M, Dunstone, M.A, Saibil, H.R. | | Deposit date: | 2014-10-20 | | Release date: | 2015-02-18 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (17 Å) | | Cite: | Conformational Changes During Pore Formation by the Perforin-Related Protein Pleurotolysin.
Plos Biol., 13, 2015
|
|
4V3N
 
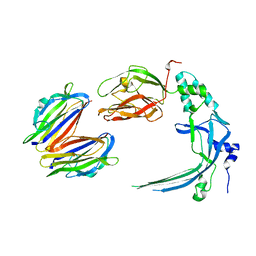 | | Membrane bound pleurotolysin prepore (TMH2 strand lock) trapped with engineered disulphide cross-link | | Descriptor: | PLEUROTOLYSIN A, PLEUROTOLYSIN B | | Authors: | Lukoyanova, N, Kondos, S.C, Farabella, I, Law, R.H.P, Reboul, C.F, Caradoc-Davies, T.T, Spicer, B.A, Kleifeld, O, Perugini, M, Ekkel, S, Hatfaludi, T, Oliver, K, Hotze, E.M, Tweten, R.K, Whisstock, J.C, Topf, M, Dunstone, M.A, Saibil, H.R. | | Deposit date: | 2014-10-20 | | Release date: | 2015-02-18 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (14 Å) | | Cite: | Conformational Changes During Pore Formation by the Perforin-Related Protein Pleurotolysin.
Plos Biol., 13, 2015
|
|
3X2Q
 
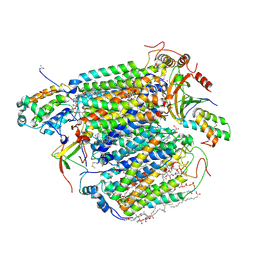 | | X-ray structure of cyanide-bound bovine heart cytochrome c oxidase in the fully oxidized state at 2.0 angstrom resolution | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Yano, N, Muramoto, K, Mochizuki, M, Shinzawa-Itoh, K, Yamashita, E, Yoshikawa, S, Tsukihara, T. | | Deposit date: | 2014-12-26 | | Release date: | 2015-06-10 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structure of cyanide-bound bovine heart cytochrome c oxidase in the fully oxidized state at 2.0 angstrom resolution.
Acta Crystallogr F Struct Biol Commun, 71, 2015
|
|
5CD7
 
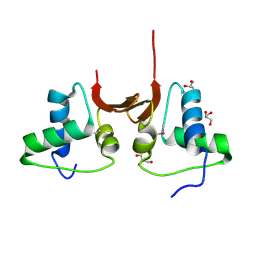 | | Crystal structure of the NTD L199M of Drosophila Oskar protein | | Descriptor: | GLYCEROL, Maternal effect protein oskar | | Authors: | Yang, N, Hu, M, Yu, Z, Wang, M, Lehmann, R, Xu, R.M. | | Deposit date: | 2015-07-03 | | Release date: | 2015-09-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Structure of Drosophila Oskar reveals a novel RNA binding protein
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5CD8
 
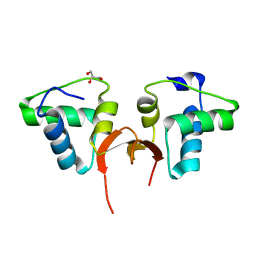 | | Crystal structure of the NTD of Drosophila Oskar protein | | Descriptor: | GLYCEROL, Maternal effect protein oskar | | Authors: | Yang, N, Hu, M, Yu, Z, Wang, M, Lehmann, R, Xu, R.M. | | Deposit date: | 2015-07-03 | | Release date: | 2015-09-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.001 Å) | | Cite: | Structure of Drosophila Oskar reveals a novel RNA binding protein
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
1JB6
 
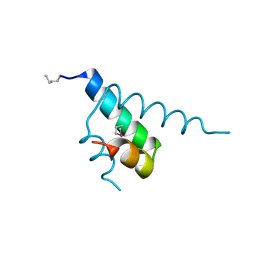 | |
5CD9
 
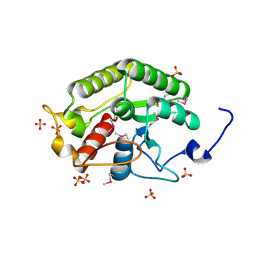 | | Crystal structure of the CTD of Drosophila Oskar protein | | Descriptor: | Maternal effect protein oskar, SULFATE ION | | Authors: | Yang, N, Hu, M, Yu, Z, Wang, M, Lehmann, R, Xu, R.M. | | Deposit date: | 2015-07-03 | | Release date: | 2015-09-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Structure of Drosophila Oskar reveals a novel RNA binding protein
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
3QYT
 
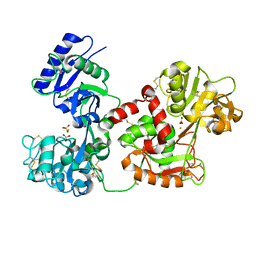 | | Diferric bound human serum transferrin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CARBONATE ION, FE (III) ION, ... | | Authors: | Yang, N, Zhang, H, Wang, M, Hao, Q, Sun, H. | | Deposit date: | 2011-03-03 | | Release date: | 2012-03-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Iron and bismuth bound human serum transferrin reveals a partially-opened conformation in the N-lobe
Sci Rep, 2, 2012
|
|
4H75
 
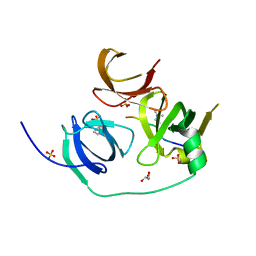 | | Crystal structure of human Spindlin1 in complex with a histone H3K4(me3) peptide | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, GLYCEROL, Histone H3, ... | | Authors: | Yang, N, Wang, W, Wang, Y, Wang, M, Zhao, Q, Rao, Z, Zhu, B, Xu, R.M. | | Deposit date: | 2012-09-20 | | Release date: | 2012-10-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.098 Å) | | Cite: | Distinct mode of methylated lysine-4 of histone H3 recognition by tandem tudor-like domains of Spindlin1.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
5B1B
 
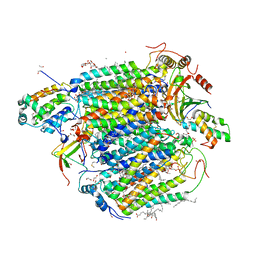 | | Bovine heart cytochrome c oxidase in the fully reduced state at 1.6 angstrom resolution | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Yano, N, Muramoto, K, Shimada, A, Takemura, S, Baba, J, Fujisawa, H, Mochizuki, M, Shinzawa-Itoh, K, Yamashita, E, Tsukihara, T, Yoshikawa, S. | | Deposit date: | 2015-12-01 | | Release date: | 2016-09-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Mg2+-containing Water Cluster of Mammalian Cytochrome c Oxidase Collects Four Pumping Proton Equivalents in Each Catalytic Cycle.
J.Biol.Chem., 291, 2016
|
|
8TCI
 
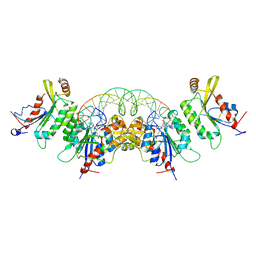 | | Crystal structure of DNMT3C-DNMT3L in complex with CGG DNA | | Descriptor: | (5'-D(P*CP*AP*TP*G)-R(P*(PYO))-D(P*GP*GP*TP*CP*TP*AP*AP*TP*TP*AP*GP*AP*CP*CP*GP*CP*AP*TP*G)-3'), DNA (cytosine-5)-methyltransferase 3-like, DNA (cytosine-5)-methyltransferase 3C, ... | | Authors: | Khudaverdyan, N, Song, J. | | Deposit date: | 2023-07-01 | | Release date: | 2024-08-14 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | The structure of DNA methyltransferase DNMT3C reveals an activity-tuning mechanism for DNA methylation.
J.Biol.Chem., 300, 2024
|
|
1BX6
 
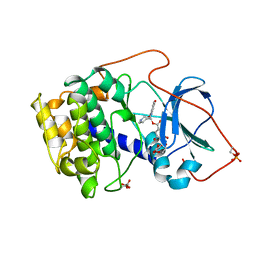 | | CRYSTAL STRUCTURE OF THE POTENT NATURAL PRODUCT INHIBITOR BALANOL IN COMPLEX WITH THE CATALYTIC SUBUNIT OF CAMP-DEPENDENT PROTEIN KINASE | | Descriptor: | BALANOL, CAMP-DEPENDENT PROTEIN KINASE | | Authors: | Narayana, N, Xuong, N.-H, Ten Eyck, L.F, Taylor, S.S. | | Deposit date: | 1998-10-13 | | Release date: | 1999-04-27 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the potent natural product inhibitor balanol in complex with the catalytic subunit of cAMP-dependent protein kinase.
Biochemistry, 38, 1999
|
|
5B1A
 
 | | Bovine heart cytochrome c oxidase in the fully oxidized state at 1.5 angstrom resolution | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Yano, N, Muramoto, K, Shimada, A, Takemura, S, Baba, J, Fujisawa, H, Mochizuki, M, Shinzawa-Itoh, K, Yamashita, E, Tsukihara, T, Yoshikawa, S. | | Deposit date: | 2015-12-01 | | Release date: | 2016-09-14 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Mg2+-containing Water Cluster of Mammalian Cytochrome c Oxidase Collects Four Pumping Proton Equivalents in Each Catalytic Cycle.
J.Biol.Chem., 291, 2016
|
|
1FMO
 
 | | CRYSTAL STRUCTURE OF A POLYHISTIDINE-TAGGED RECOMBINANT CATALYTIC SUBUNIT OF CAMP-DEPENDENT PROTEIN KINASE COMPLEXED WITH THE PEPTIDE INHIBITOR PKI(5-24) AND ADENOSINE | | Descriptor: | ADENOSINE, CAMP-DEPENDENT PROTEIN KINASE, HEAT STABLE RABBIT SKELETAL MUSCLE INHIBITOR PROTEIN | | Authors: | Narayana, N, Cox, S, Shaltiel, S, Taylor, S.S, Xuong, N.-H. | | Deposit date: | 1997-07-08 | | Release date: | 1998-01-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a polyhistidine-tagged recombinant catalytic subunit of cAMP-dependent protein kinase complexed with the peptide inhibitor PKI(5-24) and adenosine.
Biochemistry, 36, 1997
|
|
1BKX
 
 | | A BINARY COMPLEX OF THE CATALYTIC SUBUNIT OF CAMP-DEPENDENT PROTEIN KINASE AND ADENOSINE FURTHER DEFINES CONFORMATIONAL FLEXIBILITY | | Descriptor: | ADENOSINE MONOPHOSPHATE, CAMP-DEPENDENT PROTEIN KINASE | | Authors: | Narayana, N, Cox, S, Xuong, N, Ten Eyck, L.F, Taylor, S.S. | | Deposit date: | 1997-07-01 | | Release date: | 1998-03-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A binary complex of the catalytic subunit of cAMP-dependent protein kinase and adenosine further defines conformational flexibility.
Structure, 5, 1997
|
|
2ZGP
 
 | |
