1Q2R
 
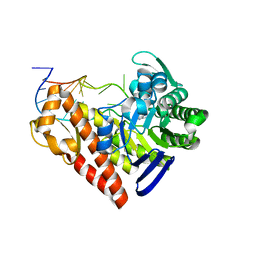 | | Chemical trapping and crystal structure of a catalytic tRNA guanine transglycosylase covalent intermediate | | Descriptor: | 9-DEAZAGUANINE, Queuine tRNA-ribosyltransferase, RNA (5'-R(*AP*GP*CP*AP*CP*GP*GP*CP*UP*(N)P*UP*AP*AP*AP*CP*CP*GP*UP*GP*C)-3'), ... | | Authors: | Xie, W, Liu, X, Huang, R.H. | | Deposit date: | 2003-07-25 | | Release date: | 2003-09-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Chemical trapping and crystal structure of a catalytic tRNA guanine transglycosylase covalent intermediate
Nat.Struct.Biol., 10, 2003
|
|
1Q2S
 
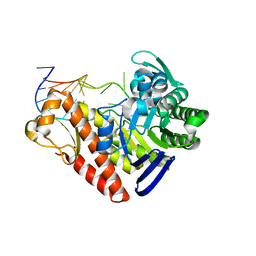 | | Chemical trapping and crystal structure of a catalytic tRNA guanine transglycosylase covalent intermediate | | Descriptor: | 5'-R(*AP*GP*CP*AP*CP*GP*GP*CP*UP*(PQ1)P*UP*AP*AP*AP*CP*CP*GP*UP*GP*C)-3', 9-DEAZAGUANINE, Queuine tRNA-ribosyltransferase, ... | | Authors: | Xie, W, Liu, X, Huang, R.H. | | Deposit date: | 2003-07-25 | | Release date: | 2003-09-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Chemical trapping and crystal structure of a catalytic tRNA guanine transglycosylase covalent intermediate
Nat.Struct.Biol., 10, 2003
|
|
6O47
 
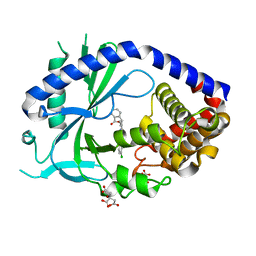 | | human cGAS core domain (K427E/K428E) bound with RU-521 | | Descriptor: | (3~{S})-3-[1-[4,5-bis(chloranyl)-1~{H}-benzimidazol-2-yl]-3-methyl-5-oxidanyl-pyrazol-4-yl]-3~{H}-2-benzofuran-1-one, 2-(4,5-dichloro-1H-benzimidazol-2-yl)-5-methyl-4-[(1R)-3-oxo-1,3-dihydro-2-benzofuran-1-yl]-1,2-dihydro-3H-pyrazol-3-one, CITRIC ACID, ... | | Authors: | Xie, W, Lama, L, Adura, C, Glickman, J.F, Tuschl, T, Patel, D.J. | | Deposit date: | 2019-02-28 | | Release date: | 2019-05-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.196 Å) | | Cite: | Human cGAS catalytic domain has an additional DNA-binding interface that enhances enzymatic activity and liquid-phase condensation.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6EDC
 
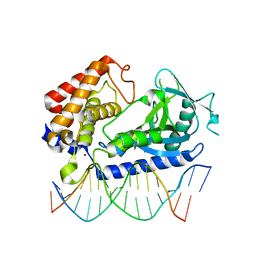 | | hcGAS-16bp dsDNA complex | | Descriptor: | Cyclic GMP-AMP synthase, DNA (5'-D(*AP*AP*AP*TP*TP*GP*CP*CP*GP*AP*AP*GP*AP*CP*GP*AP*A)-3'), DNA (5'-D(*TP*TP*CP*GP*TP*CP*TP*TP*CP*GP*GP*CP*AP*AP*T)-3'), ... | | Authors: | Xie, W, Lama, L, Adura, C, Glickman, J.F, Tuschl, T, Patel, D.J. | | Deposit date: | 2018-08-09 | | Release date: | 2019-05-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.712 Å) | | Cite: | Human cGAS catalytic domain has an additional DNA-binding interface that enhances enzymatic activity and liquid-phase condensation.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6EDB
 
 | | Crystal structure of SRY.hcGAS-21bp dsDNA complex | | Descriptor: | DNA (5'-D(*CP*GP*AP*TP*CP*CP*GP*GP*GP*AP*TP*CP*TP*AP*AP*AP*CP*AP*AP*TP*G)-3'), DNA (5'-D(*GP*CP*AP*TP*TP*GP*TP*TP*TP*AP*GP*AP*TP*CP*CP*CP*GP*GP*AP*TP*C)-3'), Sex-determining region Y protein,Cyclic GMP-AMP synthase, ... | | Authors: | Xie, W, Lama, L, Adura, C, Glickman, J.F, Tuschl, T, Patel, D.J. | | Deposit date: | 2018-08-09 | | Release date: | 2019-05-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.209 Å) | | Cite: | Human cGAS catalytic domain has an additional DNA-binding interface that enhances enzymatic activity and liquid-phase condensation.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
3JZ3
 
 | | Structure of the cytoplasmic segment of histidine kinase QseC | | Descriptor: | SULFATE ION, Sensor protein qseC | | Authors: | Xie, W, Kwiatkowski, W, Choe, S, Center for Structures of Membrane Proteins (CSMP) | | Deposit date: | 2009-09-22 | | Release date: | 2010-07-21 | | Last modified: | 2012-04-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the Cytoplasmic Segment of Histidine Kinase Receptor QseC, a Key Player in Bacterial Virulence.
Protein Pept.Lett., 17, 2010
|
|
2PMF
 
 | | The crystal structure of a human glycyl-tRNA synthetase mutant | | Descriptor: | CHLORIDE ION, GLYCEROL, Glycyl-tRNA synthetase | | Authors: | Xie, W. | | Deposit date: | 2007-04-21 | | Release date: | 2007-05-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Long-range structural effects of a Charcot-Marie- Tooth disease-causing mutation in human glycyl-tRNA synthetase.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2PME
 
 | | The Apo crystal Structure of the glycyl-tRNA synthetase | | Descriptor: | Glycyl-tRNA synthetase | | Authors: | Xie, W. | | Deposit date: | 2007-04-21 | | Release date: | 2007-05-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Long-range structural effects of a Charcot-Marie- Tooth disease-causing mutation in human glycyl-tRNA synthetase.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
6WW9
 
 | | Crystal structure of human REV7(R124A)-SHLD3(35-58) complex | | Descriptor: | Mitotic spindle assembly checkpoint protein MAD2B, Shieldin complex subunit 3 | | Authors: | Xie, W, Patel, D.J. | | Deposit date: | 2020-05-08 | | Release date: | 2021-03-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular mechanisms of assembly and TRIP13-mediated remodeling of the human Shieldin complex.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6WWA
 
 | | Crystal structure of human SHLD2-SHLD3-REV7 complex | | Descriptor: | Mitotic spindle assembly checkpoint protein MAD2B, Shieldin complex subunit 2,Shieldin complex subunit 3 chimera | | Authors: | Xie, W, Patel, D.J. | | Deposit date: | 2020-05-08 | | Release date: | 2021-03-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Molecular mechanisms of assembly and TRIP13-mediated remodeling of the human Shieldin complex.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
5CO4
 
 | |
5EGN
 
 | |
8HVJ
 
 | |
7SHO
 
 | | Crystal structure of hSTING in complex with c[2',3'-(ara-2'-G, ribo-3'-A)-MP] (RJ242) | | Descriptor: | (2R,5R,7R,8S,10R,12aR,14R,15R,15aS,16R)-7-(2-amino-6-oxo-1,6-dihydro-9H-purin-9-yl)-14-(6-amino-9H-purin-9-yl)-2,10,15,16-tetrahydroxyoctahydro-2H,10H,12H-5,8-methano-2lambda~5~,10lambda~5~-furo[3,2-l][1,3,6,9,11,2,10]pentaoxadiphosphacyclotetradecine-2,10-dione, Stimulator of interferon genes protein | | Authors: | Xie, W, Lama, L, Yang, X.J, Kuryavyi, V, Nudelman, I, Glickman, J.F, Jones, R.A, Tuschl, T, Patel, D.J. | | Deposit date: | 2021-10-10 | | Release date: | 2022-10-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Arabinose- and xylose-modified analogs of 2',3'-cGAMP act as STING agonists.
Cell Chem Biol, 2023
|
|
7SHP
 
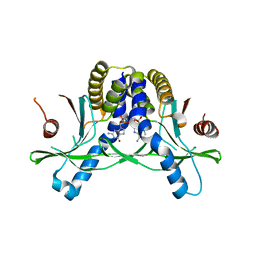 | | Crystal structure of hSTING in complex with c[2',3'-(ribo-2'-G, xylo-3'-A)-MP](RJ244) | | Descriptor: | (2S,5R,7R,8R,10S,12aR,14R,15R,15aR,16R)-7-(2-amino-6-oxo-3,6-dihydro-9H-purin-9-yl)-14-(6-amino-9H-purin-9-yl)-2,10,15,16-tetrahydroxyoctahydro-2H,10H,12H-5,8-methano-2lambda~5~,10lambda~5~-furo[3,2-l][1,3,6,9,11,2,10]pentaoxadiphosphacyclotetradecine-2,10-dione, Stimulator of interferon genes protein | | Authors: | Xie, W, Lama, L, Yang, X.J, Kuryavyi, V, Nudelman, I, Glickman, J.F, Jones, R.A, Tuschl, T, Patel, D.J. | | Deposit date: | 2021-10-11 | | Release date: | 2022-10-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Arabinose- and xylose-modified analogs of 2',3'-cGAMP act as STING agonists.
Cell Chem Biol, 2023
|
|
7JW2
 
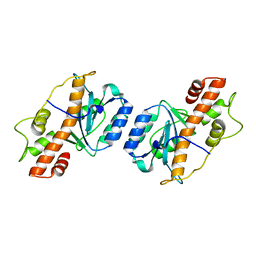 | | Crystal structure of Aedes aegypti Nibbler EXO domain | | Descriptor: | Exonuclease mut-7 homolog | | Authors: | Xie, W, Sowemimo, I, Hayashi, R, Wang, J, Brennecke, J, Ameres, S.L, Patel, D.J. | | Deposit date: | 2020-08-24 | | Release date: | 2021-01-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-function analysis of microRNA 3'-end trimming by Nibbler.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7JW6
 
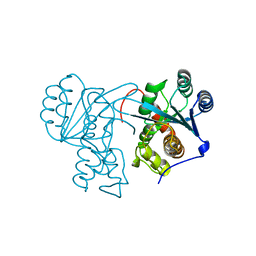 | | Crystal structure of Drosophila Nibbler EXO domain | | Descriptor: | Exonuclease mut-7 homolog | | Authors: | Xie, W, Sowemimo, I, Hayashi, R, Wang, J, Brennecke, J, Ameres, S.L, Patel, D.J. | | Deposit date: | 2020-08-24 | | Release date: | 2021-01-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-function analysis of microRNA 3'-end trimming by Nibbler.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7JW3
 
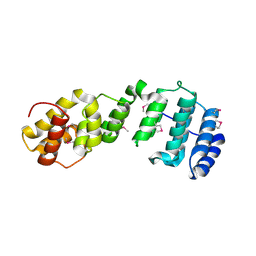 | | Crystal structure of Aedes aegypti Nibbler NTD domain | | Descriptor: | Exonuclease mut-7 homolog | | Authors: | Xie, W, Sowemimo, I, Hayashi, R, Wang, J, Brennecke, J, Ameres, S.L, Patel, D.J. | | Deposit date: | 2020-08-24 | | Release date: | 2021-01-20 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structure-function analysis of microRNA 3'-end trimming by Nibbler.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7L9P
 
 | | Structure of human SHLD2-SHLD3-REV7-TRIP13(E253Q) complex | | Descriptor: | Mitotic spindle assembly checkpoint protein MAD2B, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Pachytene checkpoint protein 2 homolog, ... | | Authors: | Xie, W, Patel, D.J. | | Deposit date: | 2021-01-04 | | Release date: | 2021-03-03 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Molecular mechanisms of assembly and TRIP13-mediated remodeling of the human Shieldin complex.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7D88
 
 | |
7D89
 
 | |
7DLC
 
 | | The structure of the Arabidopsis thaliana guanosine deaminase in reaction with N1-methylguanosine | | Descriptor: | 2-azanyl-9-[(2R,3R,4S,5R)-5-(hydroxymethyl)-3,4-bis(oxidanyl)oxolan-2-yl]-1-methyl-purin-6-one, Guanosine deaminase, ZINC ION | | Authors: | Xie, W, Jia, Q, Zeng, H. | | Deposit date: | 2020-11-27 | | Release date: | 2022-02-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Asymmetric Catalysis of Arabidopsis thaliana Guanosine Deaminase Revealed by Crystal Structures
To Be Published
|
|
7DOX
 
 | |
7DOY
 
 | | The structure of the Arabidopsis thaliana guanosine deaminase in complex with 6-O-methylguanosine | | Descriptor: | (2R,3R,4S,5R)-2-(2-azanyl-6-methoxy-purin-9-yl)-5-(hydroxymethyl)oxolane-3,4-diol, Guanosine deaminase, ZINC ION | | Authors: | Xie, W, Jia, Q, Zeng, H. | | Deposit date: | 2020-12-17 | | Release date: | 2022-02-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Asymmetric Catalysis of Arabidopsis thaliana Guanosine Deaminase Revealed by Crystal Structures
To Be Published
|
|
7DOW
 
 | | The structure of the Arabidopsis thaliana guanosine deaminase in reaction with 7-deazaguansoine | | Descriptor: | 2-azanyl-7-[(2R,3R,4S,5R)-5-(hydroxymethyl)-3,4-bis(oxidanyl)oxolan-2-yl]-3H-pyrrolo[2,3-d]pyrimidin-4-one, Guanosine deaminase, ZINC ION | | Authors: | Xie, W, Jia, Q, Zeng, H. | | Deposit date: | 2020-12-17 | | Release date: | 2022-02-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Asymmetric Catalysis of Arabidopsis thaliana Guanosine Deaminase Revealed by Crystal Structures
To Be Published
|
|
