5XVN
 
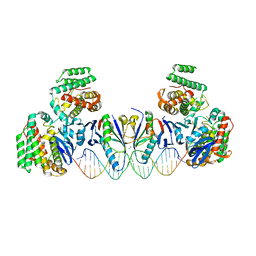 | | E. far Cas1-Cas2/prespacer binary complex | | Descriptor: | CRISPR-associated endonuclease Cas1, CRISPR-associated endoribonuclease Cas2, DNA (28-MER), ... | | Authors: | Xiao, Y, Ng, S, Nam, K.H, Ke, A. | | Deposit date: | 2017-06-28 | | Release date: | 2017-10-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | How type II CRISPR-Cas establish immunity through Cas1-Cas2-mediated spacer integration.
Nature, 550, 2017
|
|
8D6J
 
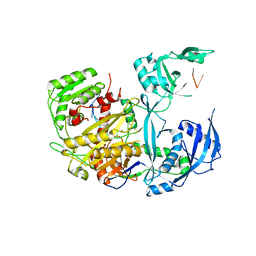 | |
8D71
 
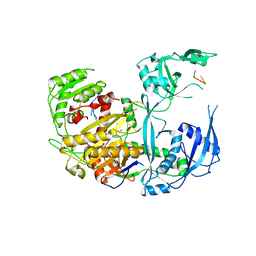 | | Human Ago2 bound to miR122(21nt) | | Descriptor: | MAGNESIUM ION, Protein argonaute-2, miR-122-21nt | | Authors: | Xiao, Y, MacRae, I. | | Deposit date: | 2022-06-07 | | Release date: | 2023-05-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A tiny loop in the Argonaute PIWI domain tunes small RNA seed strength.
Embo Rep., 24, 2023
|
|
6C66
 
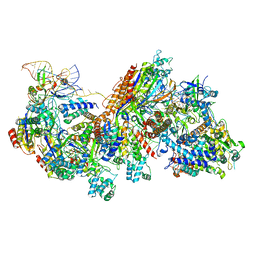 | | CRISPR RNA-guided surveillance complex, pre-nicking | | Descriptor: | CRISPR-associated helicase, Cas3 family, CRISPR-associated protein, ... | | Authors: | Xiao, Y, Luo, M, Liao, M, Ke, A. | | Deposit date: | 2018-01-17 | | Release date: | 2018-06-27 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.66 Å) | | Cite: | Structure basis for RNA-guided DNA degradation by Cascade and Cas3.
Science, 361, 2018
|
|
2AMN
 
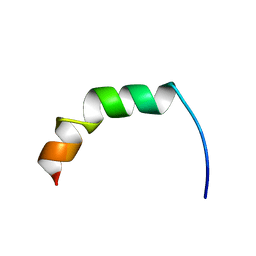 | | Solution structure of Fowlicidin-1, a novel Cathelicidin antimicrobial peptide from chicken | | Descriptor: | cathelicidin | | Authors: | Xiao, Y, Dai, H, Bommineni, Y.R, Prakash, O, Zhang, G. | | Deposit date: | 2005-08-09 | | Release date: | 2006-07-18 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Structure-activity relationships of fowlicidin-1, a cathelicidin antimicrobial peptide in chicken.
Febs J., 273, 2006
|
|
5U0A
 
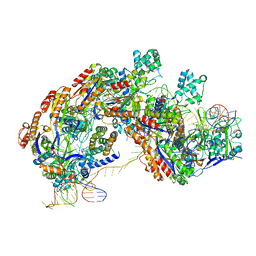 | | CRISPR RNA-guided surveillance complex | | Descriptor: | CRISPR-associated protein, Cas5e family, Cse1 family, ... | | Authors: | Xiao, Y, Luo, M, Hayes, R.P, Kim, J, Ng, S, Ding, F, Liao, M, Ke, A. | | Deposit date: | 2016-11-23 | | Release date: | 2017-08-09 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure Basis for Directional R-loop Formation and Substrate Handover Mechanisms in Type I CRISPR-Cas System.
Cell, 170, 2017
|
|
5U07
 
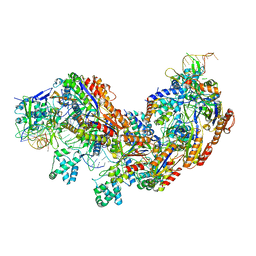 | | CRISPR RNA-guided surveillance complex | | Descriptor: | CRISPR-associated protein, Cas5e family, Cse1 family, ... | | Authors: | Xiao, Y, Luo, M, Hayes, R.P, Kim, J, Ng, S, Ding, F, Liao, M, Ke, A. | | Deposit date: | 2016-11-23 | | Release date: | 2017-08-09 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure Basis for Directional R-loop Formation and Substrate Handover Mechanisms in Type I CRISPR-Cas System.
Cell, 170, 2017
|
|
8KBD
 
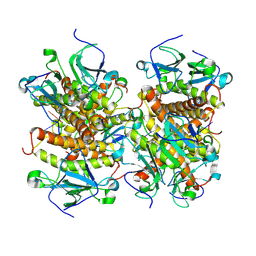 | |
8KBI
 
 | | Structure of apo-AcrIIA7 | | Descriptor: | Inhibitor of Type II CRISPR-Cas system | | Authors: | Xiao, Y, Feng, Y. | | Deposit date: | 2023-08-04 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of apo-AcrIIA7
To Be Published
|
|
8KBM
 
 | | Structure of AcrIIA7 complexed with cGG | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), Inhibitor of Type II CRISPR-Cas system | | Authors: | Xiao, Y, Feng, Y. | | Deposit date: | 2023-08-04 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Structure of AcrIIA7 complexed with cGG
To Be Published
|
|
8KBH
 
 | |
8KBJ
 
 | | Structure of HgmTad2 complexed with 1',2'-cADPR | | Descriptor: | (1S,3R,4R,6R,9S,11R,14R,15S,16R,18R)-4-(6-amino-9H-purin-9-yl)-9,11,15,16,18-pentahydroxy-2,5,8,10,12,17-hexaoxa-9lambda~5~,11lambda~5~-diphosphatricyclo[12.2.1.1~3,6~]octadecane-9,11-dione, Inhibitor of Type II CRISPR-Cas system | | Authors: | Xiao, Y, Feng, Y. | | Deposit date: | 2023-08-04 | | Release date: | 2024-08-21 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of HgmTad2 complexed with 1',2'-cADPR
To Be Published
|
|
8KBG
 
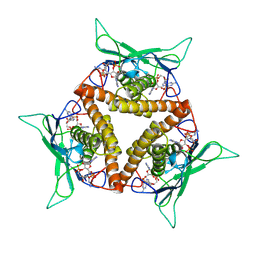 | |
8KBC
 
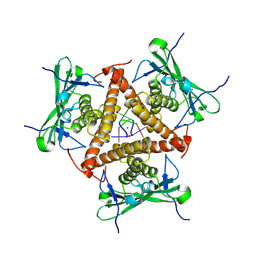 | | Structure of CmTad1 complexed with cAAA | | Descriptor: | (2-ACETYL-5-METHYLANILINO)(2,6-DIBROMOPHENYL)ACETAMIDE, Thoeris anti-defense 1, ZINC ION | | Authors: | Xiao, Y, Feng, Y. | | Deposit date: | 2023-08-04 | | Release date: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of CmTad1 complexed with cAAA
To Be Published
|
|
8KBE
 
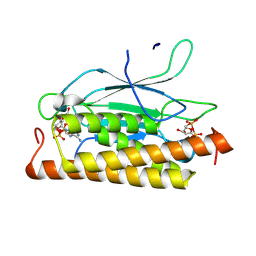 | | Structure of CbTad1 complexed with 1',3'-cADPR | | Descriptor: | (2R,3R,3aS,5S,6R,7S,8R,11R,13S,15aR)-2-(6-amino-9H-purin-9-yl)-3,6,7,11,13-pentahydroxyoctahydro-2H,5H,11H,13H-5,8-epoxy-11lambda~5~,13lambda~5~-furo[2,3-g][1,3,5,9,2,4]tetraoxadiphosphacyclotetradecine-11,13-dione, Thoeris anti-defense 1 | | Authors: | Xiao, Y, Feng, Y. | | Deposit date: | 2023-08-04 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Structure of CbTad1 complexed with 1',3'-cADPR
To Be Published
|
|
8KBF
 
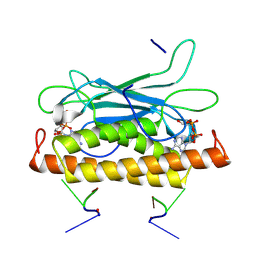 | | Structure of CbTad1 complexed with 1',3'-cADPR and cA3 | | Descriptor: | (2-ACETYL-5-METHYLANILINO)(2,6-DIBROMOPHENYL)ACETAMIDE, (2R,3R,3aS,5S,6R,7S,8R,11R,13S,15aR)-2-(6-amino-9H-purin-9-yl)-3,6,7,11,13-pentahydroxyoctahydro-2H,5H,11H,13H-5,8-epoxy-11lambda~5~,13lambda~5~-furo[2,3-g][1,3,5,9,2,4]tetraoxadiphosphacyclotetradecine-11,13-dione, Thoeris anti-defense 1 | | Authors: | Xiao, Y, Feng, Y. | | Deposit date: | 2023-08-04 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structure of CbTad1 complexed with 1',3'-cADPR and cA3
To Be Published
|
|
8KBK
 
 | | Structure of AcrIIA7 complexed with 1',2'-cADPR and cGG | | Descriptor: | (1S,3R,4R,6R,9S,11R,14R,15S,16R,18R)-4-(6-amino-9H-purin-9-yl)-9,11,15,16,18-pentahydroxy-2,5,8,10,12,17-hexaoxa-9lambda~5~,11lambda~5~-diphosphatricyclo[12.2.1.1~3,6~]octadecane-9,11-dione, 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), Inhibitor of Type II CRISPR-Cas system | | Authors: | Xiao, Y, Feng, Y. | | Deposit date: | 2023-08-04 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structure of AcrIIA7 complexed with 1',2'-cADPR and cGG
To Be Published
|
|
8KBB
 
 | | Structure of apo-CmTad1 | | Descriptor: | Thoeris anti-defense 1, ZINC ION | | Authors: | Xiao, Y, Feng, Y. | | Deposit date: | 2023-08-04 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Structure of apo-CmTad1
To Be Published
|
|
8KBL
 
 | | Structure of AcrIIA7 complexed with 1',3'-cADPR and cGG | | Descriptor: | (2R,3R,3aS,5S,6R,7S,8R,11R,13S,15aR)-2-(6-amino-9H-purin-9-yl)-3,6,7,11,13-pentahydroxyoctahydro-2H,5H,11H,13H-5,8-epoxy-11lambda~5~,13lambda~5~-furo[2,3-g][1,3,5,9,2,4]tetraoxadiphosphacyclotetradecine-11,13-dione, 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), Inhibitor of Type II CRISPR-Cas system | | Authors: | Xiao, Y, Feng, Y. | | Deposit date: | 2023-08-04 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structure of AcrIIA7 complexed with 1',3'-cADPR and cGG
To Be Published
|
|
7SWF
 
 | |
7SWQ
 
 | |
7SVA
 
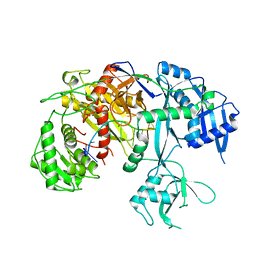 | | Cryo-EM structure of Arabidopsis Ago10-guide RNA complex | | Descriptor: | MAGNESIUM ION, Protein argonaute 10, RNA (5'-R(P*UP*GP*GP*AP*GP*UP*GP*UP*GP*AP*CP*AP*AP*UP*GP*GP*UP*GP*UP*UP*U)-3') | | Authors: | Xiao, Y, MacRae, I.J. | | Deposit date: | 2021-11-18 | | Release date: | 2022-11-23 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Structural basis for RNA slicing by a plant Argonaute.
Nat.Struct.Mol.Biol., 30, 2023
|
|
3UB0
 
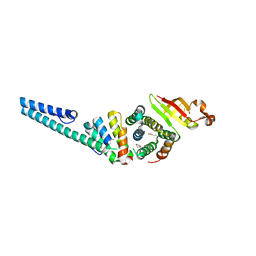 | | Crystal structure of the nonstructural protein 7 and 8 complex of Feline Coronavirus | | Descriptor: | Non-structural protein 6, nsp6,, Non-structural protein 7, ... | | Authors: | Xiao, Y, Hilgenfeld, R, Ma, Q. | | Deposit date: | 2011-10-22 | | Release date: | 2012-02-22 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Nonstructural proteins 7 and 8 of feline coronavirus form a 2:1 heterotrimer that exhibits primer-independent RNA polymerase activity.
J.Virol., 86, 2012
|
|
8H7Q
 
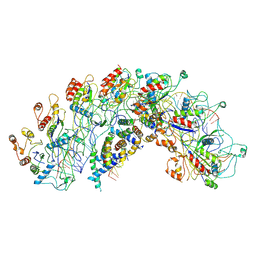 | | Cryo-EM structure of Synechocystis sp. PCC6714 Cascade at 3.8 angstrom resolution | | Descriptor: | CRISPR RNA, CRISPR associated protein Cas11b, CRISPR associated protein Cas5, ... | | Authors: | Xiao, Y, Lu, M, Yu, C, Zhang, Y. | | Deposit date: | 2022-10-20 | | Release date: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structure of Synechocystis sp. PCC6714 Cascade at 3.8 angstrom resolution
To Be Published
|
|
8IKX
 
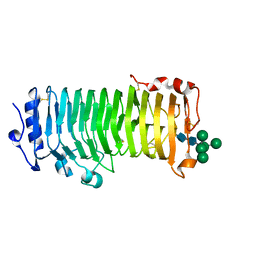 | | An Arabidopsis polygalacturonase PGLR | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Pectin lyase-like superfamily protein, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Xiao, Y, Chai, J. | | Deposit date: | 2023-03-01 | | Release date: | 2024-02-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A plant mechanism of hijacking pathogen virulence factors to trigger innate immunity.
Science, 383, 2024
|
|
