1M8W
 
 | | CRYSTAL STRUCTURE OF THE PUMILIO-HOMOLOGY DOMAIN FROM HUMAN PUMILIO1 IN COMPLEX WITH NRE1-19 RNA | | Descriptor: | 5'-R(P*UP*GP*UP*AP*UP*AP*U)-3', 5'-R(P*UP*GP*UP*CP*CP*AP*G)-3', 5'-R(P*UP*UP*GP*UP*AP*UP*AP*U)-3', ... | | Authors: | Wang, X, McLachlan, J, Zamore, P.D, Hall, T.M.T. | | Deposit date: | 2002-07-26 | | Release date: | 2002-09-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | MODULAR RECOGNITION OF RNA BY A HUMAN PUMILIO-HOMOLOGY DOMAIN
CELL(CAMBRIDGE,MASS.), 110, 2002
|
|
7P3O
 
 | |
7P3M
 
 | |
7P3P
 
 | |
1LI9
 
 | | Crystal structure of TEM-34 beta-Lactamase at 1.5 Angstrom | | Descriptor: | Class A beta-Lactamase- TEM-34, PHOSPHATE ION, POTASSIUM ION | | Authors: | Wang, X, Minasov, G, Shoichet, B.K. | | Deposit date: | 2002-04-17 | | Release date: | 2002-09-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | The structural bases of antibiotic resistance in the clinically derived mutant beta-lactamases TEM-30, TEM-32, and TEM-34.
J.Biol.Chem., 277, 2002
|
|
6NIF
 
 | | crystal structure of human REV7-RAN complex | | Descriptor: | hREV7, GTP-binding nuclear protein Ran, hREV3 fusion | | Authors: | Wang, X, Pertz, L, Hua, D.P, Zhang, T.Q, Listovsky, T, Xie, W. | | Deposit date: | 2018-12-27 | | Release date: | 2019-09-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | REV7 has a dynamic adaptor region to accommodate small GTPase RAN/ShigellaIpaB ligands, and its activity is regulated by the RanGTP/GDP switch.
J.Biol.Chem., 294, 2019
|
|
7A1V
 
 | |
5WTH
 
 | | Cryo-EM structure for Hepatitis A virus complexed with FAB | | Descriptor: | FAB Heavy Chain, FAB Light Chain, Polyprotein, ... | | Authors: | Wang, X, Zhu, L, Dang, M, Hu, Z, Gao, Q, Yuan, S, Sun, Y, Zhang, B, Ren, J, Walter, T.S, Wang, J, Fry, E.E, Stuart, D.I, Rao, Z. | | Deposit date: | 2016-12-12 | | Release date: | 2017-01-25 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Potent neutralization of hepatitis A virus reveals a receptor mimic mechanism and the receptor recognition site
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5WTF
 
 | | Cryo-EM structure for Hepatitis A virus empty particle | | Descriptor: | VP0, VP1, VP3 | | Authors: | Wang, X, Zhu, L, Dang, M, Hu, Z, Gao, Q, Yuan, S, Sun, Y, Zhang, B, Ren, J, Walter, T.S, Wang, J, Fry, E.E, Stuart, D.I, Rao, Z. | | Deposit date: | 2016-12-11 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Potent neutralization of hepatitis A virus reveals a receptor mimic mechanism and the receptor recognition site
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
2JZC
 
 | | NMR solution structure of ALG13: The sugar donor subunit of a yeast N-acetylglucosamine transferase. Northeast Structural Genomics Consortium target YG1 | | Descriptor: | UDP-N-acetylglucosamine transferase subunit ALG13 | | Authors: | Wang, X, Weldeghorghis, T, Zhang, G, Imepriali, B, Montelione, G.T, Prestegard, J.H, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-01-04 | | Release date: | 2008-02-19 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Alg13: the sugar donor subunit of a yeast N-acetylglucosamine transferase.
Structure, 16, 2008
|
|
2JT3
 
 | | Solution Structure of F153W cardiac troponin C | | Descriptor: | Troponin C | | Authors: | Wang, X, Mercier, P, Letourneau, P, Sykes, B.D. | | Deposit date: | 2007-07-18 | | Release date: | 2007-07-31 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Effects of Phe-to-Trp mutation and fluorotryptophan incorporation on the solution structure of cardiac troponin C, and analysis of its suitability as a potential probe for in situ NMR studies.
Protein Sci., 14, 2005
|
|
2JTZ
 
 | | Solution structure and chemical shift assignments of the F104-to-5-flurotryptophan mutant of cardiac troponin C | | Descriptor: | Troponin C, slow skeletal and cardiac muscles | | Authors: | Wang, X, Mercier, P, Letourneau, P, Sykes, B.D. | | Deposit date: | 2007-08-10 | | Release date: | 2007-08-28 | | Last modified: | 2021-10-20 | | Method: | SOLUTION NMR | | Cite: | Effects of Phe-to-Trp mutation and fluorotryptophan incorporation on the solution structure of cardiac troponin C, and analysis of its suitability as a potential probe for in situ NMR studies
Protein Sci., 14, 2005
|
|
2K2R
 
 | | The NMR structure of alpha-parvin CH2/paxillin LD1 complex | | Descriptor: | Alpha-parvin, Paxillin | | Authors: | Wang, X, Fukuda, K, Byeon, I, Velyvis, A, Wu, C, Gronenborn, A, Qin, J. | | Deposit date: | 2008-04-10 | | Release date: | 2008-05-27 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The Structure of {alpha}-Parvin CH2-Paxillin LD1 Complex Reveals a Novel Modular Recognition for Focal Adhesion Assembly.
J.Biol.Chem., 283, 2008
|
|
2JT0
 
 | | Solution structure of F104W cardiac troponin C | | Descriptor: | Troponin C, slow skeletal and cardiac muscles | | Authors: | Wang, X, Mercier, P, Letourneau, P.-J, Sykes, B.D. | | Deposit date: | 2007-07-17 | | Release date: | 2008-05-27 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Effects of Phe-to-Trp mutation and fluorotryptophan incorporation on the solution structure of cardiac troponin C, and analysis of its suitability as a potential probe for in situ NMR studies.
Protein Sci., 14, 2005
|
|
2JT8
 
 | | Solution structure of the F153-to-5-flurotryptophan mutant of human cardiac troponin C | | Descriptor: | Troponin C, slow skeletal and cardiac muscles | | Authors: | Wang, X, Mercier, P, Letourneau, P, Sykes, B.D. | | Deposit date: | 2007-07-20 | | Release date: | 2007-08-07 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Effects of Phe-to-Trp mutation and fluorotryptophan incorporation on the solution structure of cardiac troponin C, and analysis of its suitability as a potential probe for in situ NMR studies
Protein Sci., 14, 2005
|
|
1AKN
 
 | | STRUCTURE OF BILE-SALT ACTIVATED LIPASE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BILE-SALT ACTIVATED LIPASE | | Authors: | Wang, X, Zhang, X. | | Deposit date: | 1997-05-23 | | Release date: | 1998-05-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The crystal structure of bovine bile salt activated lipase: insights into the bile salt activation mechanism.
Structure, 5, 1997
|
|
1AQL
 
 | |
1DDJ
 
 | | CRYSTAL STRUCTURE OF HUMAN PLASMINOGEN CATALYTIC DOMAIN | | Descriptor: | PLASMINOGEN | | Authors: | Wang, X, Terzyan, S, Tang, J, Loy, J, Lin, X, Zhang, X. | | Deposit date: | 1999-11-10 | | Release date: | 2000-02-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Human plasminogen catalytic domain undergoes an unusual conformational change upon activation.
J.Mol.Biol., 295, 2000
|
|
1BML
 
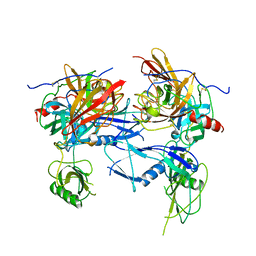 | |
8WPH
 
 | | Anabaena McyI with prebound NAD | | Descriptor: | McyI-NAD, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Wang, X, Yin, Y, Duan, Y, Liu, L. | | Deposit date: | 2023-10-10 | | Release date: | 2024-10-16 | | Last modified: | 2025-04-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural insights into the catalytic mechanism of the microcystin tailoring enzyme McyI.
Commun Biol, 8, 2025
|
|
8WPR
 
 | | Anabaena McyI R166A with prebound NAD and malate | | Descriptor: | (2S)-2-hydroxybutanedioic acid, GLYCEROL, McyI, ... | | Authors: | Wang, X, Yin, Y, Duan, Y, Liu, L. | | Deposit date: | 2023-10-10 | | Release date: | 2024-10-16 | | Last modified: | 2025-04-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insights into the catalytic mechanism of the microcystin tailoring enzyme McyI.
Commun Biol, 8, 2025
|
|
3NFC
 
 | |
4QPG
 
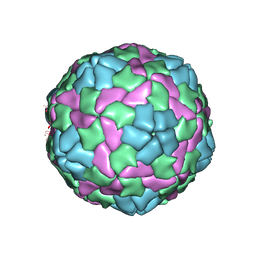 | | Crystal structure of empty hepatitis A virus | | Descriptor: | CHLORIDE ION, Capsid protein VP0, Capsid protein VP1, ... | | Authors: | Wang, X, Ren, J, Gao, Q, Hu, Z, Sun, Y, Li, X, Rowlands, D.J, Yin, W, Wang, J, Stuart, D.I, Rao, Z, Fry, E.E. | | Deposit date: | 2014-06-23 | | Release date: | 2014-10-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Hepatitis A virus and the origins of picornaviruses.
Nature, 517, 2015
|
|
4QPI
 
 | | Crystal structure of hepatitis A virus | | Descriptor: | CHLORIDE ION, Capsid protein VP1, Capsid protein VP2, ... | | Authors: | Wang, X, Ren, J, Gao, Q, Hu, Z, Sun, Y, Li, X, Rowlands, D.J, Yin, W, Wang, J, Stuart, D.I, Rao, Z, Fry, E.E. | | Deposit date: | 2014-06-23 | | Release date: | 2014-10-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Hepatitis A virus and the origins of picornaviruses.
Nature, 517, 2015
|
|
3G36
 
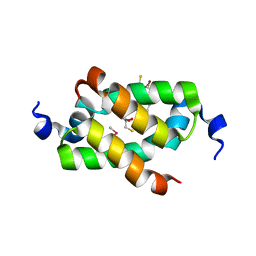 | | Crystal structure of the human DPY-30-like C-terminal domain | | Descriptor: | (2R,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, (2S,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, ... | | Authors: | Wang, X, Lou, Z, Bartlam, M, Rao, Z. | | Deposit date: | 2009-02-02 | | Release date: | 2009-06-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structure of the C-terminal domain of human DPY-30-like protein: A component of the histone methyltransferase complex
J.Mol.Biol., 390, 2009
|
|
