1QHV
 
 | | HUMAN ADENOVIRUS SEROTYPE 2 FIBRE HEAD | | Descriptor: | PROTEIN (ADENOVIRUS FIBRE), SULFATE ION | | Authors: | Van Raaij, M.J, Louis, N, Chroboczek, J, Cusack, S. | | Deposit date: | 1999-05-28 | | Release date: | 1999-09-29 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Structure of the human adenovirus serotype 2 fiber head domain at 1.5 A resolution.
Virology, 262, 1999
|
|
1QIU
 
 | |
1COW
 
 | | BOVINE MITOCHONDRIAL F1-ATPASE COMPLEXED WITH AUROVERTIN B | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, AUROVERTIN B, BOVINE MITOCHONDRIAL F1-ATPASE, ... | | Authors: | van Raaij, M.J, Abrahams, J.P, Leslie, A.G.W, Walker, J.E. | | Deposit date: | 1996-05-08 | | Release date: | 1996-08-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The structure of bovine F1-ATPase complexed with the antibiotic inhibitor aurovertin B.
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
1F5W
 
 | | DIMERIC STRUCTURE OF THE COXSACKIE VIRUS AND ADENOVIRUS RECEPTOR D1 DOMAIN | | Descriptor: | COXSACKIE VIRUS AND ADENOVIRUS RECEPTOR, SULFATE ION | | Authors: | van Raaij, M.J, Chouin, E, van der Zandt, H, Bergelson, J.M, Cusack, S. | | Deposit date: | 2000-06-18 | | Release date: | 2000-11-08 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Dimeric structure of the coxsackievirus and adenovirus receptor D1 domain at 1.7 A resolution.
Structure Fold.Des., 8, 2000
|
|
1EAJ
 
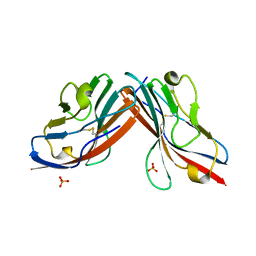 | |
5LYE
 
 | |
4UMI
 
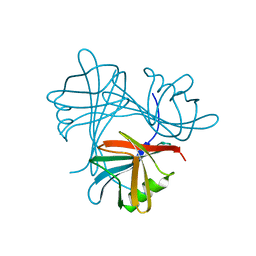 | |
4UW8
 
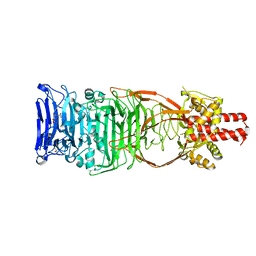 | | Structure of the carboxy-terminal domain of the bacteriophage T5 L- shaped tail fiber with its intra-molecular chaperone domain | | Descriptor: | CITRATE ANION, L-SHAPED TAIL FIBER PROTEIN | | Authors: | Garcia-Doval, C, Luque, D, Caston, J.R, Otero, J.M, Llamas-Saiz, A.L, Boulanger, P, van Raaij, M.J. | | Deposit date: | 2014-08-08 | | Release date: | 2015-08-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Structure of the Receptor-Binding Carboxy-Terminal Domain of the Bacteriophage T5 L-Shaped Tail Fibre with and without Its Intra-Molecular Chaperone.
Viruses, 7, 2015
|
|
4UE0
 
 | |
4UW7
 
 | | Structure of the carboxy-terminal domain of the bacteriophage T5 L- shaped tail fiber without its intra-molecular chaperone domain | | Descriptor: | GLYCEROL, L-SHAPED TAIL FIBER PROTEIN | | Authors: | Garcia-Doval, C, Luque, D, Caston, J.R, Otero, J.M, Llamas-Saiz, A.L, Boulanger, P, van Raaij, M.J. | | Deposit date: | 2014-08-08 | | Release date: | 2015-08-05 | | Last modified: | 2018-01-17 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Structure of the Receptor-Binding Carboxy-Terminal Domain of the Bacteriophage T5 L-Shaped Tail Fibre with and without Its Intra-Molecular Chaperone.
Viruses, 7, 2015
|
|
4UXE
 
 | | Crystal structure of the carboxy-terminal region of the bacteriophage T4 proximal long tail fibre protein gp34, P21 selenomethionine crystal | | Descriptor: | GLYCEROL, LARGE TAIL FIBER PROTEIN P34 | | Authors: | Granell, M, Alvira, S, Garcia-Doval, C, Singh, A.K, van Raaij, M.J. | | Deposit date: | 2014-08-22 | | Release date: | 2015-08-19 | | Last modified: | 2017-07-19 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Carboxy-Terminal Region of the Bacteriophage T4 Proximal Long Tail Fiber Protein Gp34.
Viruses, 9, 2017
|
|
2XGF
 
 | | Structure of the bacteriophage T4 long tail fibre needle-shaped receptor-binding tip | | Descriptor: | CARBONATE ION, FE (II) ION, LONG TAIL FIBER PROTEIN P37 | | Authors: | Bartual, S.G, Otero, J.M, Garcia-Doval, C, Llamas-Saiz, A.L, Kahn, R, Fox, G.C, van Raaij, M.J. | | Deposit date: | 2010-06-03 | | Release date: | 2010-11-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the bacteriophage T4 long tail fiber receptor-binding tip.
Proc. Natl. Acad. Sci. U.S.A., 107, 2010
|
|
4UXG
 
 | | Crystal structure of the carboxy-terminal region of the bacteriophage T4 proximal long tail fibre protein gp34, R32 native crystal | | Descriptor: | LARGE TAIL FIBER PROTEIN P34 | | Authors: | Granell, M, Alvira, S, Garcia-Doval, C, Singh, A.K, van Raaij, M.J. | | Deposit date: | 2014-08-22 | | Release date: | 2015-09-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of the Carboxy-Terminal Region of the Bacteriophage T4 Proximal Long Tail Fiber Protein Gp34.
Viruses, 9, 2017
|
|
4UXF
 
 | | Crystal structure of the carboxy-terminal region of the bacteriophage T4 proximal long tail fibre protein gp34, P21 native crystal | | Descriptor: | GLYCEROL, LARGE TAIL FIBER PROTEIN P34 | | Authors: | Granell, M, Alvira, S, Garcia-Doval, C, Singh, A.K, van Raaij, M.J. | | Deposit date: | 2014-08-22 | | Release date: | 2015-12-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Carboxy-Terminal Region of the Bacteriophage T4 Proximal Long Tail Fiber Protein Gp34.
Viruses, 9, 2017
|
|
4V0S
 
 | | Crystal structure of Mycobacterium tuberculosis Type II Dehydroquinase D88N mutant inhibited by a 3-dehydroquinic acid derivative | | Descriptor: | (1R,2S,4S,5R)-2-(2,3,4,5,6-pentafluorophenyl)methyl-1,4,5-trihydroxy-3-oxocyclohexane-1-carboxylic acid, 3,4-DIHYDROXY-2-[(2,3,4,5,6-PENTAFLUOROPHENYL)METHYL]BENZOIC ACID, 3-DEHYDROQUINATE DEHYDRATASE, ... | | Authors: | Otero, J.M, Llamas-Saiz, A.L, Santiago, C, Lamb, H, Hawkins, A.R, Maneiro, M, Peon, A, Gonzalez-Bello, C, van Raaij, M.J. | | Deposit date: | 2014-09-18 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Investigation of the Dehydratation Mechanism Catalyzed by the Type II Dehydroquinase
To be Published
|
|
2XB8
 
 | | Structure of Mycobacterium tuberculosis type II dehydroquinase in complex with inhibitor compound (2R)-2-(4-methoxybenzyl)-3- dehydroquinic acid | | Descriptor: | (1R,2R,4S,5R)-1,4,5-TRIHYDROXY-2-(4-METHOXYBENZYL)-3-OXOCYCLOHEXANECARBOXYLIC ACID, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-DEHYDROQUINATE DEHYDRATASE, ... | | Authors: | Otero, J.M, Tizon, L, Llamas-Saiz, A.L, Fox, G.C, Gonzalez-Bello, C, van Raaij, M.J. | | Deposit date: | 2010-04-08 | | Release date: | 2010-09-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Understanding the Key Factors that Control the Inhibition of Type II Dehydroquinase by (2R)-2- Benzyl-3-Dehydroquinic Acids.
Chemmedchem, 5, 2010
|
|
4D0V
 
 | | Crystal structure of the fiber head domain of the Atadenovirus snake adenovirus 1, native, I213 crystal form | | Descriptor: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, FIBER PROTEIN, SULFATE ION | | Authors: | Singh, A.K, van Raaij, M.J. | | Deposit date: | 2014-04-30 | | Release date: | 2014-12-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the fibre head domain of the Atadenovirus Snake Adenovirus 1.
PLoS ONE, 9, 2014
|
|
4CSH
 
 | | Native structure of the lytic CHAPK domain of the endolysin LysK from Staphylococcus aureus bacteriophage K | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GLYCEROL, ... | | Authors: | Sanz-Gaitero, M, Keary, R, Garcia-Doval, C, Coffey, A, van Raaij, M.J. | | Deposit date: | 2014-03-07 | | Release date: | 2014-08-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Crystal structure of the lytic CHAP(K) domain of the endolysin LysK from Staphylococcus aureus bacteriophage K.
Virol. J., 11, 2014
|
|
4D1F
 
 | |
4D62
 
 | |
4CIY
 
 | | Crystal structure of Mycobacterium tuberculosis type 2 dehydroquinase in complex with (1R,4R,5R)-1,4,5-trihydroxy-3-((1R)-1-hydroxy-2- phenyl)ethylcyclohex-2-en-1-carboxylic acid | | Descriptor: | (1R,4R,5R)-1,4,5-trihydroxy-3-[(1R)-1-hydroxy-2-phenyl]ethylcyclohex-2-ene-1-carboxylic acid, 3-DEHYDROQUINATE DEHYDRATASE, CHLORIDE ION, ... | | Authors: | Otero, J.M, Llamas-Saiz, A.L, Lamb, H, Hawkins, A.R, Blanco, B, Sedes, A, Peon, A, Gonzalez-Bello, C, van Raaij, M.J. | | Deposit date: | 2013-12-17 | | Release date: | 2014-04-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Exploring the water-binding pocket of the type II dehydroquinase enzyme in the structure-based design of inhibitors.
J. Med. Chem., 57, 2014
|
|
4CIW
 
 | | Crystal structure of Mycobacterium tuberculosis type 2 dehydroquinase in complex with (1R,4R,5R)-1,4,5-trihydroxy-3-(2-hydroxy)ethylcyclohex-2-ene-1-carboxylic acid | | Descriptor: | (1R,4R,5R)-1,4,5-trihydroxy-3-(2-hydroxy)ethylcyclohex-2-ene-1-carboxylic acid, 3-DEHYDROQUINATE DEHYDRATASE, SODIUM ION, ... | | Authors: | Otero, J.M, Llamas-Saiz, A.L, Lamb, H, Hawkins, A.R, Blanco, B, Sedes, A, Peon, A, Gonzalez-Bello, C, van Raaij, M.J. | | Deposit date: | 2013-12-17 | | Release date: | 2014-04-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Exploring the water-binding pocket of the type II dehydroquinase enzyme in the structure-based design of inhibitors.
J. Med. Chem., 57, 2014
|
|
4CIV
 
 | | Crystal structure of Mycobacterium tuberculosis type 2 dehydroquinase in complex with (1R,4R,5R)-1,4,5-trihydroxy-3-hydroxymethylcyclohex-2-ene-1-carboxylic acid | | Descriptor: | (1R,4R,5R)-1,4,5-trihydroxy-3-hydroxymethylcyclohex-2-ene-1-carboxylic acid, 3-DEHYDROQUINATE DEHYDRATASE | | Authors: | Otero, J.M, Llamas-Saiz, A.L, Lamb, H, Hawkins, A.R, Blanco, B, Sedes, A, Peon, A, Gonzalez-Bello, C, van Raaij, M.J. | | Deposit date: | 2013-12-17 | | Release date: | 2014-04-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Exploring the water-binding pocket of the type II dehydroquinase enzyme in the structure-based design of inhibitors.
J. Med. Chem., 57, 2014
|
|
4CL0
 
 | | Structure of the Mycobacterium tuberculosis Type II Dehydroquinase inhibited by a 3-dehydroquinic acid derivative | | Descriptor: | (2R)-2-METHYL-3-DEHYDROQUINIC ACID, 3-DEHYDROQUINATE DEHYDRATASE | | Authors: | Otero, J.M, Llamas-Saiz, A.L, Maneiro, M, Peon, A, Sedes, A, Lamb, H, Hawkins, A.R, Gonzalez-Bello, C, van Raaij, M.J. | | Deposit date: | 2014-01-10 | | Release date: | 2015-03-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Investigation of the Dehydratation Mechanism Catalyzed by the Type II Dehydroquinase
To be Published
|
|
4CKW
 
 | | Structure of the Mycobacterium tuberculosis Type II Dehydroquinase N12S mutant (Crystal Form 1) | | Descriptor: | 3-DEHYDROQUINATE DEHYDRATASE, GLYCEROL | | Authors: | Otero, J.M, Llamas-Saiz, A.L, Maneiro, M, Peon, A, Sedes, A, Lamb, H, Hawkins, A.R, Gonzalez-Bello, C, van Raaij, M.J. | | Deposit date: | 2014-01-10 | | Release date: | 2015-03-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Investigation of the Dehydratation Mechanism Catalyzed by the Type II Dehydroquinase
To be Published
|
|
