9FZW
 
 | | Structure of Urethanase UMG-SP2 | | Descriptor: | GLYCEROL, SULFATE ION, Urethanase UMG-SP2 | | Authors: | Singh, P, Lennartz, F, Bornscheuer, U.T, Wei, R, Weber, G. | | Deposit date: | 2024-07-06 | | Release date: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Structure of Urethanase UMG-SP2
To Be Published
|
|
5DUI
 
 | |
3C4W
 
 | |
3C4Z
 
 | |
3C51
 
 | |
3C4X
 
 | |
3C50
 
 | |
3C4Y
 
 | |
2XI1
 
 | | Crystal structure of the HIV-1 Nef sequenced from a patient's sample | | Descriptor: | NEF | | Authors: | Yadav, G.P, Singh, P, Gupta, S, Tripathi, A.K, Tripathi, R.K, Ramachandran, R. | | Deposit date: | 2010-06-25 | | Release date: | 2011-08-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | A Novel Dimer-Tetramer Transition Captured by the Crystal Structure of the HIV-1 Nef.
Plos One, 6, 2011
|
|
1X3E
 
 | | Crystal structure of the single-stranded DNA-binding protein from Mycobacterium smegmatis | | Descriptor: | CADMIUM ION, Single-strand binding protein | | Authors: | Saikrishnan, K, Manjunath, G.P, Singh, P, Jeyakanthan, J, Dauter, Z, Sekar, K, Muniyappa, K, Vijayan, M. | | Deposit date: | 2005-05-04 | | Release date: | 2005-08-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of Mycobacterium smegmatis single-stranded DNA-binding protein and a comparative study involving homologus SSBs: biological implications of structural plasticity and variability in quaternary association.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1X3G
 
 | | Crystal structure of the single-stranded DNA-binding protein from Mycobacterium SMEGMATIS | | Descriptor: | CADMIUM ION, Single-strand binding protein | | Authors: | Saikrishnan, K, Manjunath, G.P, Singh, P, Jeyakanthan, J, Dauter, Z, Sekar, K, Muniyappa, K, Vijayan, M. | | Deposit date: | 2005-05-05 | | Release date: | 2005-08-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of Mycobacterium smegmatis single-stranded DNA-binding protein and a comparative study involving homologus SSBs: biological implications of structural plasticity and variability in quaternary association.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1X3F
 
 | | Crystal structure of the single-stranded DNA-binding protein from Mycobacterium SMEGMATIS | | Descriptor: | CADMIUM ION, Single-strand binding protein | | Authors: | Saikrishnan, K, Manjunath, G.P, Singh, P, Jeyakanthan, J, Dauter, Z, Sekar, K, Muniyappa, K, Vijayan, M. | | Deposit date: | 2005-05-05 | | Release date: | 2005-08-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of Mycobacterium smegmatis single-stranded DNA-binding protein and a comparative study involving homologus SSBs: biological implications of structural plasticity and variability in quaternary association.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
6CHT
 
 | | HNF4alpha in complex with the corepressor EBP1 fragment | | Descriptor: | Hepatocyte nuclear factor 4-alpha, LAURIC ACID, Proliferation-associated protein 2G4 | | Authors: | Chi, Y.I, Singh, P, Lee, I.K. | | Deposit date: | 2018-02-22 | | Release date: | 2019-02-27 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (3.174 Å) | | Cite: | ErbB3-binding protein 1 (EBP1) represses HNF4 alpha-mediated transcription and insulin secretion in pancreatic beta-cells.
J.Biol.Chem., 294, 2019
|
|
3AFP
 
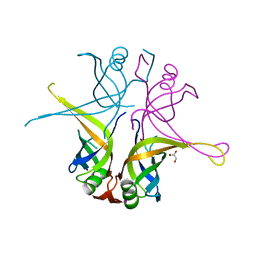 | | Crystal structure of the single-stranded DNA binding protein from Mycobacterium leprae (Form I) | | Descriptor: | CADMIUM ION, GLYCEROL, Single-stranded DNA-binding protein | | Authors: | Kaushal, P.S, Singh, P, Sharma, A, Muniyappa, K, Vijayan, M. | | Deposit date: | 2010-03-10 | | Release date: | 2010-10-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | X-ray and molecular-dynamics studies on Mycobacterium leprae single-stranded DNA-binding protein and comparison with other eubacterial SSB structures
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3AFQ
 
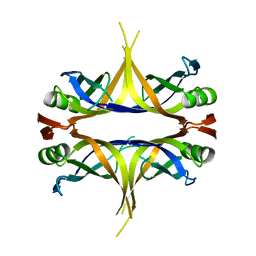 | | Crystal structure of the single-stranded DNA binding protein from Mycobacterium leprae (Form II) | | Descriptor: | Single-stranded DNA-binding protein | | Authors: | Kaushal, P.S, Singh, P, Sharma, A, Muniyappa, K, Vijayan, M. | | Deposit date: | 2010-03-10 | | Release date: | 2010-10-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-ray and molecular-dynamics studies on Mycobacterium leprae single-stranded DNA-binding protein and comparison with other eubacterial SSB structures
Acta Crystallogr.,Sect.D, 66, 2010
|
|
4E1Q
 
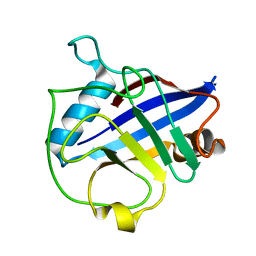 | | Crystal structure of Wheat Cyclophilin A at 1.25 A resolution | | Descriptor: | Peptidyl-prolyl cis-trans isomerase | | Authors: | Sekhon, S.S, Jeong, D.G, Woo, E.J, Singh, P, Yoon, T.S. | | Deposit date: | 2012-03-06 | | Release date: | 2013-03-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.251 Å) | | Cite: | Structural and biochemical characterization of the cytosolic wheat cyclophilin TaCypA-1
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4HY7
 
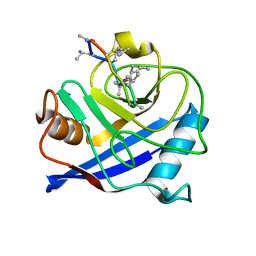 | | Structural and biochemical characterization of a cytosolic wheat cyclophilin TaCypA-1 | | Descriptor: | Cyclosporin A, Peptidyl-prolyl cis-trans isomerase | | Authors: | Sekhon, S.S, Jeong, D.G, Woo, E.J, Singh, P, Pareek, A, Yoon, T.-S. | | Deposit date: | 2012-11-13 | | Release date: | 2013-03-27 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural and biochemical characterization of the cytosolic wheat cyclophilin TaCypA-1.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3NYN
 
 | | Crystal Structure of G Protein-Coupled Receptor Kinase 6 in Complex with Sangivamycin | | Descriptor: | (R,R)-2,3-BUTANEDIOL, G protein-coupled receptor kinase 6, SANGIVAMYCIN, ... | | Authors: | Tesmer, J.J.G, Singh, P. | | Deposit date: | 2010-07-15 | | Release date: | 2010-09-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Molecular basis for activation of G protein-coupled receptor kinases.
Embo J., 29, 2010
|
|
3T8O
 
 | | Rhodopsin kinase (GRK1) L166K mutant at 2.5A resolution | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Tesmer, J.J.G, Singh, P, Nance, M.R. | | Deposit date: | 2011-08-01 | | Release date: | 2012-06-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of a monomeric variant of rhodopsin kinase at 2.5 A resolution.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
1ODZ
 
 | | Expansion of the glycosynthase repertoire to produce defined manno-oligosaccharides | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Mannan endo-1,4-beta-mannosidase, SODIUM ION, ... | | Authors: | Jahn, M, Stoll, D, Warren, R.A.J, Szabo, L, Singh, P, Gilbert, H.J, Ducros, V.M.A, Davies, G.J, Withers, S.G. | | Deposit date: | 2003-03-17 | | Release date: | 2003-07-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Expansion of the Glycosynthase Repertoire to Produce Defined Manno-Oligosaccharides
Chem.Commun.(Camb.), 12, 2003
|
|
3NYO
 
 | | Crystal Structure of G Protein-Coupled Receptor Kinase 6 in Complex with AMP | | Descriptor: | (R,R)-2,3-BUTANEDIOL, ADENOSINE MONOPHOSPHATE, G protein-coupled receptor kinase 6, ... | | Authors: | Tesmer, J.J.G, Singh, P. | | Deposit date: | 2010-07-15 | | Release date: | 2010-09-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Molecular basis for activation of G protein-coupled receptor kinases.
Embo J., 29, 2010
|
|
3S37
 
 | |
3S34
 
 | | Structure of the 1121B Fab fragment | | Descriptor: | 1121B Fab heavy chain, 1121B Fab light chain, PHOSPHATE ION | | Authors: | Franklin, M.C. | | Deposit date: | 2011-05-17 | | Release date: | 2011-08-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Structural Basis for the Function of Two Anti-VEGF Receptor 2 Antibodies.
Structure, 19, 2011
|
|
8TDN
 
 | |
8TDO
 
 | |
