1C28
 
 | |
1NCH
 
 | | STRUCTURAL BASIS OF CELL-CELL ADHESION BY CADHERINS | | Descriptor: | N-CADHERIN, YTTERBIUM (III) ION | | Authors: | Shapiro, L, Fannon, A.M, Kwong, P.D, Thompson, A, Lehmann, M.S, Grubel, G, Legrand, J.-F, Als-Nielsen, J, Colman, D.R, Hendrickson, W.A. | | Deposit date: | 1995-03-23 | | Release date: | 1995-07-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of cell-cell adhesion by cadherins.
Nature, 374, 1995
|
|
1NCG
 
 | | STRUCTURAL BASIS OF CELL-CELL ADHESION BY CADHERINS | | Descriptor: | N-CADHERIN, YTTERBIUM (III) ION | | Authors: | Shapiro, L, Fannon, A.M, Kwong, P.D, Thompson, A, Lehmann, M.S, Grubel, G, Legrand, J.-F, Als-Nielsen, J, Colman, D.R, Hendrickson, W.A. | | Deposit date: | 1995-03-23 | | Release date: | 1995-07-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of cell-cell adhesion by cadherins.
Nature, 374, 1995
|
|
1NCI
 
 | | STRUCTURAL BASIS OF CELL-CELL ADHESION BY CADHERINS | | Descriptor: | N-CADHERIN, URANYL (VI) ION | | Authors: | Shapiro, L, Fannon, A.M, Kwong, P.D, Thompson, A, Lehmann, M.S, Grubel, G, Legrand, J.-F, Als-Nielsen, J, Colman, D.R, Hendrickson, W.A. | | Deposit date: | 1995-03-23 | | Release date: | 1995-07-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of cell-cell adhesion by cadherins.
Nature, 374, 1995
|
|
1NEU
 
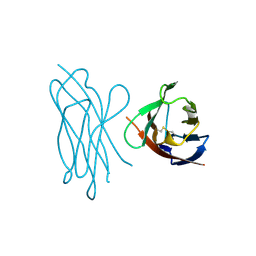 | | STRUCTURE OF MYELIN MEMBRANE ADHESION MOLECULE P0 | | Descriptor: | MYELIN P0 PROTEIN | | Authors: | Shapiro, L, Doyle, J.P, Hensley, P, Colman, D.R, Hendrickson, W.A. | | Deposit date: | 1996-09-24 | | Release date: | 1997-05-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the extracellular domain from P0, the major structural protein of peripheral nerve myelin.
Neuron, 17, 1996
|
|
1C3H
 
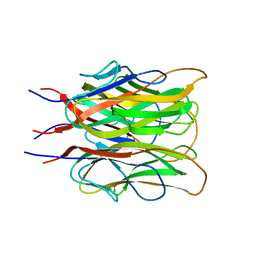 | | ACRP30 CALCIUM COMPLEX | | Descriptor: | 30 KD ADIPOCYTE COMPLEMENT-RELATED PROTEIN PRECURSOR, CALCIUM ION | | Authors: | Shapiro, L, Boggon, T, Scherer, P. | | Deposit date: | 1999-07-27 | | Release date: | 2003-12-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | ACRP30 calcium complex
To Be Published
|
|
6WUD
 
 | |
6WU7
 
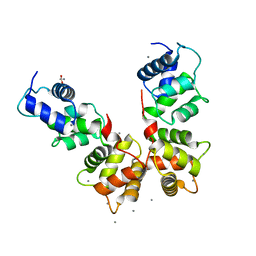 | |
3K6I
 
 | |
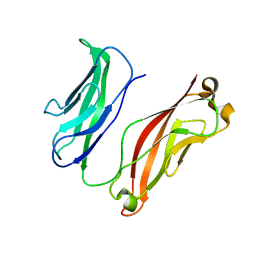
3K5R
 
 | |
3K6D
 
 | |
3K6F
 
 | |
3K5S
 
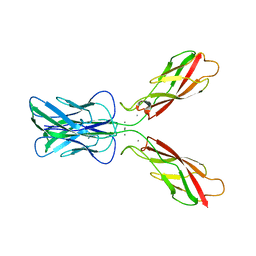 | |
6EFZ
 
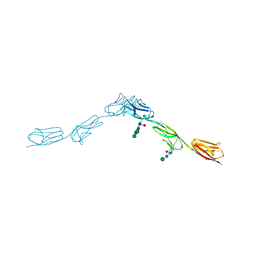 | | Crystal Structure of DIP-Theta Ig1-3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Cosmanescu, F, Shapiro, L. | | Deposit date: | 2018-08-17 | | Release date: | 2018-11-28 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.499 Å) | | Cite: | Neuron-Subtype-Specific Expression, Interaction Affinities, and Specificity Determinants of DIP/Dpr Cell Recognition Proteins.
Neuron, 100, 2018
|
|
7RW2
 
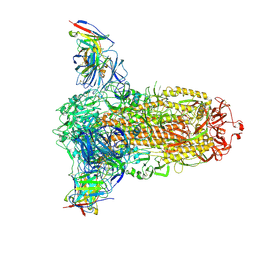 | |
7SD5
 
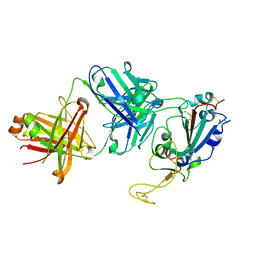 | | Crystallographic structure of neutralizing antibody 10-40 in complex with SARS-CoV-2 spike receptor binding domain | | Descriptor: | 10-40 Heavy chain, 10-40 Light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Reddem, E.R, Casner, R.G, Shapiro, L. | | Deposit date: | 2021-09-29 | | Release date: | 2022-04-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | An antibody class with a common CDRH3 motif broadly neutralizes sarbecoviruses.
Sci Transl Med, 14, 2022
|
|
7SI2
 
 | |
1Q7E
 
 | | Crystal Structure of YfdW protein from E. coli | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Hypothetical protein yfdW, METHIONINE | | Authors: | Gogos, A, Gorman, J, Shapiro, L, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2003-08-18 | | Release date: | 2003-09-30 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of Escherichia coli YfdW, a type III CoA transferase.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
5IRY
 
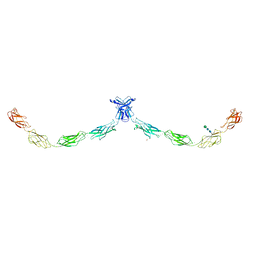 | | Crystal structure of human Desmocollin-1 ectodomain | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Brasch, J, Harrison, O.J, Shapiro, L. | | Deposit date: | 2016-03-15 | | Release date: | 2016-06-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.095 Å) | | Cite: | Structural basis of adhesive binding by desmocollins and desmogleins.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
7UKL
 
 | |
2A4E
 
 | | Crystal structure of mouse cadherin-11 EC1-2 | | Descriptor: | CALCIUM ION, Cadherin-11 | | Authors: | Patel, S.D, Ciatto, C, Chen, C.P, Bahna, F, Arkus, N, Rajebhosale, M, Jessell, T.M, Honig, B, Price, S.R, Shapiro, L. | | Deposit date: | 2005-06-28 | | Release date: | 2006-04-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Type II cadherin ectodomain structures: implications for classical cadherin specificity.
Cell(Cambridge,Mass.), 124, 2006
|
|
1ZWK
 
 | |
1ZXK
 
 | | Crystal Structure of Cadherin8 EC1 domain | | Descriptor: | Cadherin-8 | | Authors: | Patel, S.D, Ciatto, C, Chen, C.P, Bahna, F, Arkus, N, Schieren, I, Rajebhosale, M, Jessell, T.M, Honig, B, Price, S.R, Shapiro, L. | | Deposit date: | 2005-06-08 | | Release date: | 2006-04-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Type II cadherin ectodomain structures: implications for classical cadherin specificity.
Cell(Cambridge,Mass.), 124, 2006
|
|
2A1F
 
 | |
2A4C
 
 | | Crystal structure of mouse cadherin-11 EC1 | | Descriptor: | Cadherin-11 | | Authors: | Patel, S.D, Ciatto, C, Chen, C.P, Bahna, F, Schieren, I, Rajebhosale, M, Jessell, T.M, Honig, B, Price, S.R, Shapiro, L. | | Deposit date: | 2005-06-28 | | Release date: | 2006-04-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Type II cadherin ectodomain structures: implications for classical cadherin specificity.
Cell(Cambridge,Mass.), 124, 2006
|
|
