2D94
 
 | | THE CONFORMATION OF THE DNA DOUBLE HELIX IN THE CRYSTAL IS DEPENDENT ON ITS ENVIRONMENT | | Descriptor: | DNA (5'-D(*GP*GP*GP*CP*GP*CP*CP*C)-3') | | Authors: | Shakked, Z, Guerstein-Guzikevich, G, Eisenstein, M, Frolow, F, Rabinovich, D. | | Deposit date: | 1993-07-13 | | Release date: | 1994-01-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The conformation of the DNA double helix in the crystal is dependent on its environment.
Nature, 342, 1989
|
|
7KCI
 
 | | DETERMINANTS OF REPRESSOR/OPERATOR RECOGNITION FROM THE STRUCTURE OF THE TRP OPERATOR BINDING SITE | | Descriptor: | Self-complementary deoxyoligonucleotide decamer d(CCACTAGTGG) | | Authors: | Shakked, Z, Guzikevich-Guerstein, G, Frolow, F, Rabinovich, D, Joachimiak, A, Sigler, P.B. | | Deposit date: | 1994-09-12 | | Release date: | 2020-10-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Determinants of repressor/operator recognition from the structure of the trp operator binding site.
Nature, 368, 1994
|
|
1VJ4
 
 | | SEQUENCE-DEPENDENT CONFORMATION OF AN A-DNA DOUBLE HELIX: THE CRYSTAL STRUCTURE OF THE OCTAMER D(G-G-T-A-T-A-C-C) | | Descriptor: | 5'-D(*GP*GP*TP*AP*TP*AP*CP*C)-3' | | Authors: | Shakked, Z, Rabinovich, D, Kennard, O, Cruse, W.B, Salisbury, S.A, Viswamitra, M.A. | | Deposit date: | 1989-01-11 | | Release date: | 1989-01-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Sequence-dependent conformation of an A-DNA double helix: The crystal structure of the octamer d(G-G-T-A-T-A-C-C)
J.Mol.Biol., 166, 1983
|
|
1VT8
 
 | | Crystal structure of D(GGGCGCCC)-hexagonal form | | Descriptor: | DNA (5'-D(*GP*GP*GP*CP*GP*CP*CP*C)-3') | | Authors: | Shakked, Z, Guerstein-Guzikevich, G, Eisenstein, M, Frolow, F, Rabinovich, D. | | Deposit date: | 1996-12-02 | | Release date: | 2011-07-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The conformation of the DNA double helix in the crystal is dependent on its environment.
Nature, 342, 1989
|
|
3KZ8
 
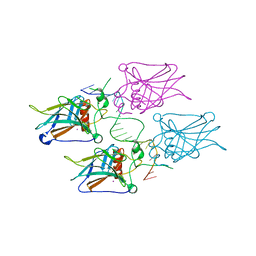 | | Diversity in DNA recognition by p53 revealed by crystal structures with Hoogsteen base pairs (p53-DNA complex 3) | | Descriptor: | Cellular tumor antigen p53, DNA (5'-D(*TP*GP*GP*GP*CP*AP*TP*GP*CP*CP*CP*GP*GP*GP*CP*AP*TP*GP*CP*CP*C)-3'), IODIDE ION, ... | | Authors: | Rozenberg, H, Suad, O, Shakked, Z. | | Deposit date: | 2009-12-08 | | Release date: | 2010-03-31 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Diversity in DNA recognition by p53 revealed by crystal structures with Hoogsteen base pairs
Nat.Struct.Mol.Biol., 17, 2010
|
|
115D
 
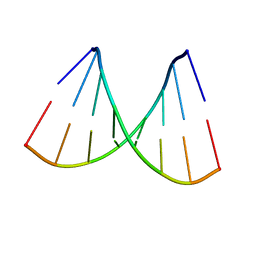 | | ORDERED WATER STRUCTURE IN AN A-DNA OCTAMER AT 1.7 ANGSTROMS RESOLUTION | | Descriptor: | DNA (5'-D(*GP*GP*(BRU)P*AP*(BRU)P*AP*CP*C)-3') | | Authors: | Kennard, O, Cruse, W.B.T, Nachman, J, Prange, T, Shakked, Z, Rabinovich, D. | | Deposit date: | 1993-02-12 | | Release date: | 1993-07-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Ordered water structure in an A-DNA octamer at 1.7 A resolution.
J.Biomol.Struct.Dyn., 3, 1986
|
|
6ZNC
 
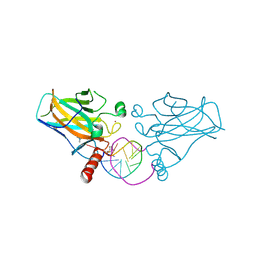 | | Structural basis of reactivation of oncogenic p53 mutants by a small molecule: methylene quinuclidinone (MQ). Human wild-type p53DBD bound to DNA and MQ: wt-DNA-MQ (I) | | Descriptor: | (2~{R})-2-methyl-1-azabicyclo[2.2.2]octan-3-one, (2~{S})-2-methyl-1-azabicyclo[2.2.2]octan-3-one, Cellular tumor antigen p53, ... | | Authors: | Rozenberg, H, Degtjarik, O, Diskin-Posner, Y, Shakked, Z. | | Deposit date: | 2020-07-06 | | Release date: | 2021-12-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structural basis of reactivation of oncogenic p53 mutants by a small molecule: methylene quinuclidinone (MQ).
Nat Commun, 12, 2021
|
|
2D95
 
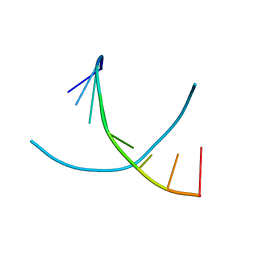 | | LOW-TEMPERATURE STUDY OF THE A-DNA FRAGMENT D(GGGCGCCC) | | Descriptor: | DNA (5'-D(*GP*GP*GP*CP*GP*CP*CP*C)-3') | | Authors: | Eisenstein, M, Hope, H, Haran, T.E, Frolow, F, Shakked, Z, Rabinovich, D. | | Deposit date: | 1993-07-13 | | Release date: | 1994-01-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Low-temperature study of the A-DNA fragment d(GGGCGCCC)
ACTA CRYSTALLOGR.,SECT.B, 44, 1988
|
|
5MG7
 
 | | New Insights into the Role of DNA Shape on Its Recognition by p53 Proteins (complex p53DBD-p53R2) | | Descriptor: | Cellular tumor antigen p53, DNA, ZINC ION | | Authors: | Rozenberg, H, Braeuning, B, Golovenko, D, Shakked, Z. | | Deposit date: | 2016-11-21 | | Release date: | 2018-06-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | New Insights into the Role of DNA Shape on Its Recognition by p53 Proteins.
Structure, 26, 2018
|
|
5MCU
 
 | | New Insights into the Role of DNA Shape on Its Recognition by p53 Proteins (complex p53DBD-LHG2) | | Descriptor: | 1,2-ETHANEDIOL, Cellular tumor antigen p53, DNA, ... | | Authors: | Golovenko, D, Rozenberg, H, Shakked, Z. | | Deposit date: | 2016-11-10 | | Release date: | 2018-06-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | New Insights into the Role of DNA Shape on Its Recognition by p53 Proteins.
Structure, 26, 2018
|
|
5MCV
 
 | | New Insights into the Role of DNA Shape on Its Recognition by p53 Proteins (complex p53DBD-LWC1) | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Cellular tumor antigen p53, ... | | Authors: | Golovenko, D, Rozenberg, H, Shakked, Z. | | Deposit date: | 2016-11-10 | | Release date: | 2018-06-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | New Insights into the Role of DNA Shape on Its Recognition by p53 Proteins.
Structure, 26, 2018
|
|
5MCW
 
 | | New Insights into the Role of DNA Shape on Its Recognition by p53 Proteins (complex p53DBD-LWC2) | | Descriptor: | Cellular tumor antigen p53, DNA, FORMYL GROUP, ... | | Authors: | Golovenko, D, Rozenberg, H, Shakked, Z. | | Deposit date: | 2016-11-10 | | Release date: | 2018-06-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.897 Å) | | Cite: | New Insights into the Role of DNA Shape on Its Recognition by p53 Proteins.
Structure, 26, 2018
|
|
188D
 
 | |
5MF7
 
 | | New Insights into the Role of DNA Shape on Its Recognition by p53 Proteins (complex p53DBD-GADD45) | | Descriptor: | Cellular tumor antigen p53, DI(HYDROXYETHYL)ETHER, DNA, ... | | Authors: | Rozenberg, H, Diskin-Posner, Y, Golovenko, D, Shakked, Z. | | Deposit date: | 2016-11-17 | | Release date: | 2018-05-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | New Insights into the Role of DNA Shape on Its Recognition by p53 Proteins.
Structure, 26, 2018
|
|
187D
 
 | |
5MCT
 
 | | New Insights into the Role of DNA Shape on Its Recognition by p53 Proteins (complex p53DBD-LHG1) | | Descriptor: | 1,2-ETHANEDIOL, Cellular tumor antigen p53, DNA, ... | | Authors: | Golovenko, D, Rozenberg, H, Shakked, Z. | | Deposit date: | 2016-11-10 | | Release date: | 2018-06-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.446 Å) | | Cite: | New Insights into the Role of DNA Shape on Its Recognition by p53 Proteins.
Structure, 26, 2018
|
|
1VTR
 
 | | STRUCTURE OF THE DEOXYTETRANUCLEOTIDE D-PAPTPAPT AND A SEQUENCE-DEPENDENT MODEL FOR POLY(DA-DT) | | Descriptor: | DNA (5'-D(*AP*TP*AP*T)-3') | | Authors: | Viswamitra, M.A, Shakked, Z, Jones, P.G, Sheldrick, G.M, Salisbury, S.A, Kennard, O. | | Deposit date: | 1988-08-18 | | Release date: | 2011-07-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Structure of the Deoxytetranucleotide d-pApTpApT and a Sequence-Dependent Model for Poly(dA-dT)
Biopolymers, 21, 1982
|
|
1VTA
 
 | |
6FJ5
 
 | | New Insights into the Role of DNA Shape on Its Recognition by p53 Proteins (complex p53DBD-AGG-HG) | | Descriptor: | 1,2-ETHANEDIOL, Cellular tumor antigen p53, DNA, ... | | Authors: | Golovenko, D, Rozenberg, H, Shakked, Z. | | Deposit date: | 2018-01-20 | | Release date: | 2018-06-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.051 Å) | | Cite: | New Insights into the Role of DNA Shape on Its Recognition by p53 Proteins.
Structure, 26, 2018
|
|
6GJC
 
 | | Structure of Mycobacterium tuberculosis Fatty Acid Synthase - I | | Descriptor: | FLAVIN MONONUCLEOTIDE, Fatty acid synthase | | Authors: | Elad, N, Baron, S, Shakked, Z, Zimhony, O, Diskin, R. | | Deposit date: | 2018-05-16 | | Release date: | 2018-09-05 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of Type-I Mycobacterium tuberculosis fatty acid synthase at 3.3 angstrom resolution.
Nat Commun, 9, 2018
|
|
287D
 
 | |
2ADY
 
 | | Structural Basis of DNA Recognition by p53 Tetramers (complex IV) | | Descriptor: | 5'-D(*CP*GP*GP*AP*CP*AP*TP*GP*TP*CP*CP*G)-3', Cellular tumor antigen p53, ZINC ION | | Authors: | Kitayner, M, Rozenberg, H, Kessler, N, Rabinovich, D, Shakked, Z. | | Deposit date: | 2005-07-21 | | Release date: | 2006-07-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis of DNA Recognition by p53 Tetramers
Mol.Cell, 22, 2006
|
|
2AC0
 
 | | Structural Basis of DNA Recognition by p53 Tetramers (complex I) | | Descriptor: | 5'-D(*CP*GP*GP*GP*CP*AP*TP*GP*CP*CP*CP*G)-3', Cellular tumor antigen p53, ZINC ION | | Authors: | Kitayner, M, Rozenberg, H, Kessler, N, Rabinovich, D, Shakked, Z. | | Deposit date: | 2005-07-18 | | Release date: | 2006-07-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis of DNA Recognition by p53 Tetramers
Mol.Cell, 22, 2006
|
|
286D
 
 | |
285D
 
 | |
