1QGO
 
 | | ANAEROBIC COBALT CHELATASE IN COBALAMIN BIOSYNTHESIS FROM SALMONELLA TYPHIMURIUM | | Descriptor: | ANAEROBIC COBALAMIN BIOSYNTHETIC COBALT CHELATASE, SULFATE ION | | Authors: | Schubert, H.L, Raux, E, Warren, M.J, Wilson, K.S. | | Deposit date: | 1999-05-03 | | Release date: | 1999-09-29 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Common chelatase design in the branched tetrapyrrole pathways of heme and anaerobic cobalamin synthesis.
Biochemistry, 38, 1999
|
|
1CBF
 
 | | THE X-RAY STRUCTURE OF A COBALAMIN BIOSYNTHETIC ENZYME, COBALT PRECORRIN-4 METHYLTRANSFERASE, CBIF | | Descriptor: | COBALT-PRECORRIN-4 TRANSMETHYLASE, PHOSPHATE ION, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Schubert, H.L, Raux, E, Woodcock, S.C, Wilson, K.S, Warren, M.J. | | Deposit date: | 1998-05-01 | | Release date: | 1999-05-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The X-ray structure of a cobalamin biosynthetic enzyme, cobalt-precorrin-4 methyltransferase.
Nat.Struct.Biol., 5, 1998
|
|
4I6M
 
 | | Structure of Arp7-Arp9-Snf2(HSA)-RTT102 subcomplex of SWI/SNF chromatin remodeler. | | Descriptor: | Actin-like protein ARP9, Actin-related protein 7, PHOSPHATE ION, ... | | Authors: | Schubert, H.L, Cairns, B.R, Hill, C.P. | | Deposit date: | 2012-11-29 | | Release date: | 2013-02-13 | | Last modified: | 2017-08-16 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | Structure of an actin-related subcomplex of the SWI/SNF chromatin remodeler.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
2CBF
 
 | | THE X-RAY STRUCTURE OF A COBALAMIN BIOSYNTHETIC ENZYME, COBALT PRECORRIN-4 METHYLTRANSFERASE, CBIF, FROM BACILLUS MEGATERIUM, WITH THE HIS-TAG CLEAVED OFF | | Descriptor: | COBALT-PRECORRIN-4 TRANSMETHYLASE, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Schubert, H.L, Raux, E, Woodcock, S.C, Warren, M.J, Wilson, K.S. | | Deposit date: | 1998-05-01 | | Release date: | 1999-05-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The X-ray structure of a cobalamin biosynthetic enzyme, cobalt-precorrin-4 methyltransferase.
Nat.Struct.Biol., 5, 1998
|
|
1YTS
 
 | | A LIGAND-INDUCED CONFORMATIONAL CHANGE IN THE YERSINIA PROTEIN TYROSINE PHOSPHATASE | | Descriptor: | SULFATE ION, YERSINIA PROTEIN TYROSINE PHOSPHATASE | | Authors: | Schubert, H.L, Stuckey, J.A, Fauman, E.B, Dixon, J.E, Saper, M.A. | | Deposit date: | 1995-04-07 | | Release date: | 1995-07-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A ligand-induced conformational change in the Yersinia protein tyrosine phosphatase.
Protein Sci., 4, 1995
|
|
1KYQ
 
 | | Met8p: A bifunctional NAD-dependent dehydrogenase and ferrochelatase involved in siroheme synthesis. | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Siroheme biosynthesis protein MET8 | | Authors: | Schubert, H.L, Raux, E, Brindley, A.A, Wilson, K.S, Hill, C.P, Warren, M.J. | | Deposit date: | 2002-02-05 | | Release date: | 2002-05-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structure of Saccharomyces cerevisiae Met8p, a bifunctional dehydrogenase and ferrochelatase.
EMBO J., 21, 2002
|
|
1NV8
 
 | | N5-glutamine methyltransferase, HemK | | Descriptor: | N5-METHYLGLUTAMINE, S-ADENOSYLMETHIONINE, hemK protein | | Authors: | Schubert, H.L, Phillips, J.D, Hill, C.P. | | Deposit date: | 2003-02-02 | | Release date: | 2003-05-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures along the Catalytic Pathway of PrmC/HemK, an N(5)-Glutamine
AdoMet-Dependent Methyltransferase
Biochemistry, 42, 2003
|
|
1NV9
 
 | | HemK, apo structure | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, hemK protein | | Authors: | Schubert, H.L, Phillips, J.D, Hill, C.P. | | Deposit date: | 2003-02-02 | | Release date: | 2003-05-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.356 Å) | | Cite: | Structures along the Catalytic Pathway of PrmC/HemK, an N(5)-Glutamine
AdoMet-Dependent Methyltransferase
Biochemistry, 42, 2003
|
|
3NWH
 
 | | Crystal structure of BST2/Tetherin | | Descriptor: | Bone marrow stromal antigen 2 | | Authors: | Schubert, H.L, Zhai, Q, Hill, C.P. | | Deposit date: | 2010-07-09 | | Release date: | 2010-07-21 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and functional studies on the extracellular domain of BST2/tetherin in reduced and oxidized conformations.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3D8N
 
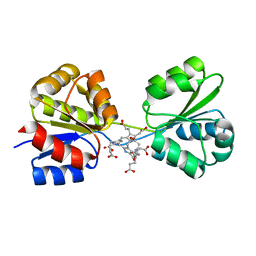 | | Uroporphyrinogen III Synthase-Uroporphyringen III Complex | | Descriptor: | 3,3',3'',3'''-[3,8,13,17-tetrakis(carboxymethyl)porphyrin-2,7,12,18-tetrayl]tetrapropanoic acid, Uroporphyrinogen-III synthase | | Authors: | Schubert, H.L. | | Deposit date: | 2008-05-23 | | Release date: | 2008-08-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and mechanistic implications of a uroporphyrinogen III synthase-product complex.
Biochemistry, 47, 2008
|
|
3D8T
 
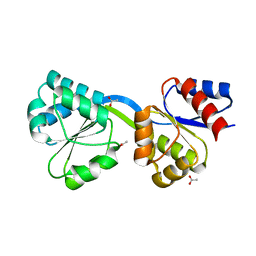 | | Thermus thermophilus Uroporphyrinogen III Synthase | | Descriptor: | ACETATE ION, Uroporphyrinogen-III synthase | | Authors: | Schubert, H.L. | | Deposit date: | 2008-05-23 | | Release date: | 2008-08-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure and mechanistic implications of a uroporphyrinogen III synthase-product complex.
Biochemistry, 47, 2008
|
|
3D8S
 
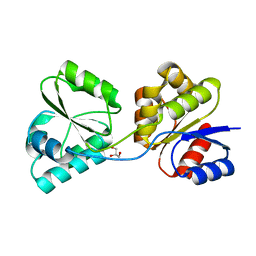 | | Thermus thermophilus Uroporphyrinogen III Synthase | | Descriptor: | GLYCEROL, Uroporphyrinogen-III synthase | | Authors: | Schubert, H.L. | | Deposit date: | 2008-05-23 | | Release date: | 2008-08-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and mechanistic implications of a uroporphyrinogen III synthase-product complex.
Biochemistry, 47, 2008
|
|
3DFZ
 
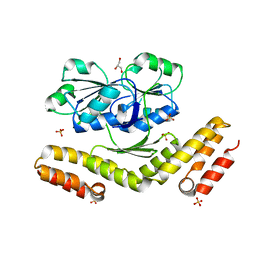 | | SirC, precorrin-2 dehydrogenase | | Descriptor: | GLYCEROL, Precorrin-2 dehydrogenase, SULFATE ION | | Authors: | Schubert, H.L, Hill, C.P, Warren, M.J. | | Deposit date: | 2008-06-12 | | Release date: | 2008-10-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and function of SirC from Bacillus megaterium: a metal-binding precorrin-2 dehydrogenase
Biochem.J., 415, 2008
|
|
3D8R
 
 | | Thermus thermophilus Uroporphyrinogen III Synthase | | Descriptor: | PHOSPHATE ION, Uroporphyrinogen-III synthase | | Authors: | Schubert, H.L. | | Deposit date: | 2008-05-23 | | Release date: | 2008-08-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and mechanistic implications of a uroporphyrinogen III synthase-product complex.
Biochemistry, 47, 2008
|
|
2IDX
 
 | |
3FRT
 
 | |
3FRR
 
 | |
3FRS
 
 | | Structure of human IST1(NTD) (residues 1-189)(p43212) | | Descriptor: | GLYCEROL, Uncharacterized protein KIAA0174 | | Authors: | Schubert, H.L, Hill, C.P, Bajorek, M, Sundquist, W.I. | | Deposit date: | 2009-01-08 | | Release date: | 2009-06-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structural basis for ESCRT-III protein autoinhibition.
Nat.Struct.Mol.Biol., 16, 2009
|
|
2XU6
 
 | | MDV1 coiled coil domain | | Descriptor: | MDV1 COILED COIL | | Authors: | Koirala, S, Bui, H.T, Schubert, H.L, Eckert, D.M, Hill, C.P, Kay, M.S, Shaw, J.M. | | Deposit date: | 2010-10-14 | | Release date: | 2010-10-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular Architecture of a Dynamin Adaptor: Implications for Assembly of Mitochondrial Fission Complexes
J.Cell Biol., 191, 2010
|
|
5WHG
 
 | | Vms1 mitochondrial localization core | | Descriptor: | Protein VMS1, ZINC ION | | Authors: | Fredrickson, E.K, Schubert, H.L, Rutter, J, Hill, C.P. | | Deposit date: | 2017-07-17 | | Release date: | 2017-11-15 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Sterol Oxidation Mediates Stress-Responsive Vms1 Translocation to Mitochondria.
Mol. Cell, 68, 2017
|
|
4N02
 
 | | Type 2 IDI from S. pneumoniae | | Descriptor: | 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, Isopentenyl-diphosphate delta-isomerase, SULFATE ION | | Authors: | de Ruyck, J, Schubert, H.L, Poulter, C.D. | | Deposit date: | 2013-09-30 | | Release date: | 2014-07-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Determination of Kinetics and the Crystal Structure of a Novel Type 2 Isopentenyl Diphosphate: Dimethylallyl Diphosphate Isomerase from Streptococcus pneumoniae.
Chembiochem, 15, 2014
|
|
6PEK
 
 | | Structure of Spastin Hexamer (Subunit A-E) in complex with substrate peptide | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ... | | Authors: | Han, H, Schubert, H.L, McCullough, J, Monroe, N, Sundquist, W.I, Hill, C.P. | | Deposit date: | 2019-06-20 | | Release date: | 2019-12-04 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structure of spastin bound to a glutamate-rich peptide implies a hand-over-hand mechanism of substrate translocation.
J.Biol.Chem., 295, 2020
|
|
6PEN
 
 | | Structure of Spastin Hexamer (whole model) in complex with substrate peptide | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, EYEYEYEYEY, ... | | Authors: | Han, H, Schubert, H.L, McCullough, J, Monroe, N, Sundquist, W.I, Hill, C.P. | | Deposit date: | 2019-06-20 | | Release date: | 2019-12-04 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structure of spastin bound to a glutamate-rich peptide implies a hand-over-hand mechanism of substrate translocation.
J.Biol.Chem., 295, 2020
|
|
1YTW
 
 | | YERSINIA PTPASE COMPLEXED WITH TUNGSTATE | | Descriptor: | SULFATE ION, TUNGSTATE(VI)ION, YERSINIA PROTEIN TYROSINE PHOSPHATASE | | Authors: | Fauman, E.B, Schubert, H.L, Saper, M.A. | | Deposit date: | 1996-05-01 | | Release date: | 1996-11-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The X-ray crystal structures of Yersinia tyrosine phosphatase with bound tungstate and nitrate. Mechanistic implications.
J.Biol.Chem., 271, 1996
|
|
3LL9
 
 | |
