7TLH
 
 | | Murine meteorin C-terminal NTR domain | | Descriptor: | Meteorin, SULFATE ION | | Authors: | Quan, C, Arndt, J.W. | | Deposit date: | 2022-01-18 | | Release date: | 2023-01-25 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Identification and structure determination of stable domains of meteorin and meteorin-like
To Be Published
|
|
7TLW
 
 | | Murine Meteorin-like C-terminal NTR domain | | Descriptor: | IODIDE ION, Meteorin-like protein | | Authors: | Quan, C, Arndt, J.W. | | Deposit date: | 2022-01-19 | | Release date: | 2023-01-25 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Identification and structure determination of stable domains of meteorin and meteorin-like
To Be Published
|
|
7TL6
 
 | | Murine meteorin N-terminal CUB domain | | Descriptor: | Meteorin, SULFATE ION | | Authors: | Quan, C, Arndt, J.W, Pepinsky, B, Gong, B.J, Dolnikova, J. | | Deposit date: | 2022-01-18 | | Release date: | 2023-01-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification and structure determination of stable domains of meteorin and meteorin-like
To Be Published
|
|
5K8D
 
 | | Crystal structure of rFVIIIFc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, COPPER (II) ION, ... | | Authors: | Leksa, N, Quan, C. | | Deposit date: | 2016-05-29 | | Release date: | 2017-06-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (4.19 Å) | | Cite: | The structural basis for the functional comparability of factor VIII and the long-acting variant recombinant factor VIII Fc fusion protein.
J. Thromb. Haemost., 15, 2017
|
|
3IAB
 
 | | Crystal structure of RNase P /RNase MRP proteins Pop6, Pop7 in a complex with the P3 domain of RNase MRP RNA | | Descriptor: | P3 domain of the RNA component of RNase MRP, Ribonucleases P/MRP protein subunit POP6, Ribonucleases P/MRP protein subunit POP7, ... | | Authors: | Perederina, A, Esakova, O.A, Quan, C, Khanova, E, Krasilnikov, A.S. | | Deposit date: | 2009-07-13 | | Release date: | 2010-01-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Eukaryotic ribonucleases P/MRP: the crystal structure of the P3 domain
Embo J., 29, 2010
|
|
6PXR
 
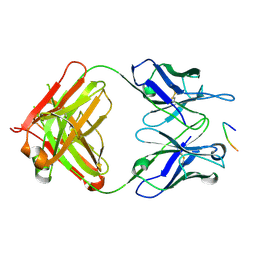 | | Anti-TAU BIIB092 FAB with TAU peptide | | Descriptor: | Microtubule-associated protein tau, gosuranemab Fab, heavy chain, ... | | Authors: | Arndt, J.W, Quan, C. | | Deposit date: | 2019-07-26 | | Release date: | 2020-07-29 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.556 Å) | | Cite: | Characterization of tau binding by gosuranemab.
Neurobiol.Dis., 146, 2020
|
|
1ILQ
 
 | |
1ILP
 
 | |
1IN3
 
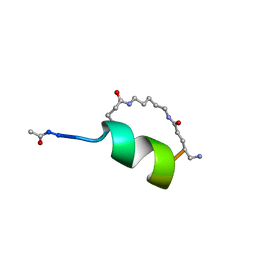 | | Peptide Antagonist of IGFBP1, (i,i+8) Covalently Restrained Analog | | Descriptor: | IGFBP-1 antagonist, PENTANE | | Authors: | Skelton, N.J, Chen, Y.M, Dubree, N, Quan, C, Jackson, D.Y, Cochran, A.G, Zobel, K, Deshayes, K, Baca, M, Pisabarro, M.T, Lowman, H.B. | | Deposit date: | 2001-05-11 | | Release date: | 2001-05-30 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Structure-function analysis of a phage display-derived peptide that binds to insulin-like growth factor binding protein 1.
Biochemistry, 40, 2001
|
|
1IN2
 
 | | Peptide Antagonist of IGFBP1, (i,i+7) Covalently Restrained Analog | | Descriptor: | IGFBP-1 antagonist, PENTANE | | Authors: | Skelton, N.J, Chen, Y.M, Dubree, N, Quan, C, Jackson, D.Y, Cochran, A.G, Zobel, K, Deshayes, K, Baca, M, Pisabarro, M.T, Lowman, H.B. | | Deposit date: | 2001-05-11 | | Release date: | 2001-05-30 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Structure-function analysis of a phage display-derived peptide that binds to insulin-like growth factor binding protein 1.
Biochemistry, 40, 2001
|
|
1GJG
 
 | | Peptide Antagonist of IGFBP1, (i,i+8) Covalently Restrained Analog, Minimized Average Structure | | Descriptor: | IGFBP-1 antagonist, PENTANE | | Authors: | Skelton, N.J, Chen, Y.M, Dubree, N, Quan, C, Jackson, D.Y, Cochran, A.G, Zobel, K, Deshayes, K, Baca, M, Pisabarro, M.T, Lowman, H.B. | | Deposit date: | 2001-05-11 | | Release date: | 2001-05-30 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure-function analysis of a phage display-derived peptide that binds to insulin-like growth factor binding protein 1.
Biochemistry, 40, 2001
|
|
1GJF
 
 | | Peptide Antagonist of IGFBP1, (i,i+7) Covalently Restrained Analog, Minimized Average Structure | | Descriptor: | IGFBP-1 antagonist, PENTANE | | Authors: | Skelton, N.J, Chen, Y.M, Dubree, N, Quan, C, Jackson, D.Y, Cochran, A.G, Zobel, K, Deshayes, K, Baca, M, Pisabarro, M.T, Lowman, H.B. | | Deposit date: | 2001-05-11 | | Release date: | 2001-05-30 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure-function analysis of a phage display-derived peptide that binds to insulin-like growth factor binding protein 1.
Biochemistry, 40, 2001
|
|
4OQT
 
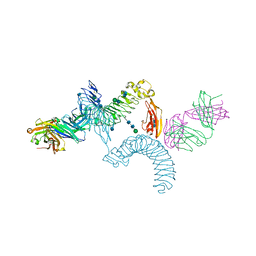 | | LINGO-1/Li81 Fab complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy Chain of Li81 Fab, ... | | Authors: | Pepinsky, R.B, Arndt, J.W, Quan, C, Gao, Y, Quintero-Monzon, O, Lee, X, Mi, S. | | Deposit date: | 2014-02-10 | | Release date: | 2014-05-14 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.23 Å) | | Cite: | Structure of the LINGO-1-Anti-LINGO-1 Li81 Antibody Complex Provides Insights into the Biology of LINGO-1 and the Mechanism of Action of the Antibody Therapy.
J.Pharmacol.Exp.Ther., 350, 2014
|
|
2WVC
 
 | | Structural and mechanistic insights into Helicobacter pylori NikR function | | Descriptor: | FORMIC ACID, GLYCEROL, PUTATIVE NICKEL-RESPONSIVE REGULATOR, ... | | Authors: | Dian, C, Bahlawane, C, Muller, C, Round, A, Delay, C, Fauquant, C, Schauer, K, de Reuse, H, Michaud-Soret, I, Terradot, L. | | Deposit date: | 2009-10-16 | | Release date: | 2010-01-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and Mechanistic Insights Into Helicobacter Pylori Nikr Activation.
Nucleic Acids Res., 38, 2010
|
|
2WVB
 
 | | Structural and mechanistic insights into Helicobacter pylori NikR function | | Descriptor: | FORMIC ACID, GLYCEROL, PUTATIVE NICKEL-RESPONSIVE REGULATOR | | Authors: | Dian, C, Bahlawane, C, Muller, C, Round, A, Delay, C, Fauquant, C, Schauer, K, De Reuse, H, Michaud-Soret, I, Terradot, L. | | Deposit date: | 2009-10-16 | | Release date: | 2010-01-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and Mechanistic Insights Into Helicobacter Pylori Nikr Activation.
Nucleic Acids Res., 38, 2010
|
|
2WVD
 
 | | Structural and mechanistic insights into Helicobacter pylori NikR function | | Descriptor: | GLYCEROL, PUTATIVE NICKEL-RESPONSIVE REGULATOR, SULFATE ION | | Authors: | Dian, C, Bahlawane, C, Muller, C, Round, A, Delay, C, Fauquant, C, Schauer, K, de Reuse, H, Michaud-Soret, I, Terradot, L. | | Deposit date: | 2009-10-16 | | Release date: | 2010-01-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural and Mechanistic Insights Into Helicobacter Pylori Nikr Activation.
Nucleic Acids Res., 38, 2010
|
|
2WVE
 
 | | Structural and mechanistic insights into Helicobacter pylori NikR function | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CITRIC ACID, GLYCEROL, ... | | Authors: | Dian, C, Bahlawane, C, Muller, C, Round, A, Delay, C, Fauquant, C, Schauer, K, de Reuse, H, Michaud-Soret, I, Terradot, L. | | Deposit date: | 2009-10-16 | | Release date: | 2010-01-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Mechanistic Insights Into Helicobacter Pylori Nikr Activation.
Nucleic Acids Res., 38, 2010
|
|
2WVF
 
 | | Structural and mechanistic insights into Helicobacter pylori NikR function | | Descriptor: | FORMIC ACID, GLYCEROL, NICKEL (II) ION, ... | | Authors: | Dian, C, Bahlawane, C, Muller, C, Round, A, Delay, C, Fauquant, C, Schauer, K, de Reuse, H, Michaud-Soret, I, Terradot, L. | | Deposit date: | 2009-10-16 | | Release date: | 2010-01-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and Mechanistic Insights Into Helicobacter Pylori Nikr Activation.
Nucleic Acids Res., 38, 2010
|
|
5WXC
 
 | | Crystal Structure of HLA-A*2402 in complex with avian influenza A(H7N9) virus-derived peptide H7-25 (data set 2) | | Descriptor: | Beta-2-microglobulin, H7-25-F, HLA class I histocompatibility antigen, ... | | Authors: | Zhao, M, Liu, K, Chai, Y, Qi, J, Liu, J, Gao, G.F. | | Deposit date: | 2017-01-07 | | Release date: | 2018-01-17 | | Last modified: | 2019-07-31 | | Method: | X-RAY DIFFRACTION (2.295 Å) | | Cite: | Heterosubtypic Protections against Human-Infecting Avian Influenza Viruses Correlate to Biased Cross-T-Cell Responses.
Mbio, 9, 2018
|
|
5WXD
 
 | | Crystal Structure of HLA-A*2402 in complex with avian influenza A(H7N9) virus-derived peptide H7-25 (data set 1) | | Descriptor: | Beta-2-microglobulin, H7-25, HLA class I histocompatibility antigen, ... | | Authors: | Zhao, M, Liu, K, Chai, Y, Qi, J, Liu, J, Gao, G.F. | | Deposit date: | 2017-01-07 | | Release date: | 2018-01-17 | | Last modified: | 2019-07-31 | | Method: | X-RAY DIFFRACTION (3.295 Å) | | Cite: | Heterosubtypic Protections against Human-Infecting Avian Influenza Viruses Correlate to Biased Cross-T-Cell Responses.
Mbio, 9, 2018
|
|
6RLX
 
 | |
4EJF
 
 | |
2YQ7
 
 | | Structure of Bcl-xL bound to BimLOCK | | Descriptor: | BCL-2-LIKE PROTEIN 1, BCL-2-LIKE PROTEIN 11, GLYCEROL | | Authors: | Smith, B.J, Czabotar, P.E. | | Deposit date: | 2012-11-06 | | Release date: | 2012-11-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Stabilizing the Pro-Apoptotic Bimbh3 Helix (Bimsahb) Does not Necessarily Enhance Affinity or Biological Activity.
Acs Chem.Biol., 8, 2013
|
|
2YQ6
 
 | | Structure of Bcl-xL bound to BimSAHB | | Descriptor: | BCL-2-LIKE PROTEIN 1, BIM BETA 5, GLYCEROL | | Authors: | Smith, B.J, Czabotar, P.E. | | Deposit date: | 2012-11-06 | | Release date: | 2012-11-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.799 Å) | | Cite: | Stabilizing the Pro-Apoptotic Bimbh3 Helix (Bimsahb) Does not Necessarily Enhance Affinity or Biological Activity.
Acs Chem.Biol., 8, 2013
|
|
3K48
 
 | | Crystal structure of APRIL bound to a peptide | | Descriptor: | Tumor necrosis factor ligand superfamily member 13, peptide | | Authors: | Hymowitz, S.G. | | Deposit date: | 2009-10-05 | | Release date: | 2009-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Multiple novel classes of APRIL-specific receptor-blocking peptides isolated by phage display.
J.Mol.Biol., 396, 2010
|
|
