1RYK
 
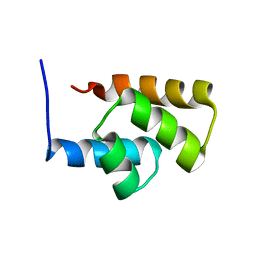 | | Solution NMR Structure Protein yjbJ from Escherichia coli. Northeast Structural Genomics Consortium Target ET93; Ontario Centre for Structural Proteomics target EC0298_1_69; | | Descriptor: | Protein yjbJ | | Authors: | Pineda-Lucena, A, Liao, J, Wu, B, Yee, A, Cort, J.R, Kennedy, M.A, Edwards, A.M, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-12-22 | | Release date: | 2004-02-24 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | An NMR approach to structural proteomics.
Proteins, 47, 2002
|
|
1N6Z
 
 | |
1MV3
 
 | |
1MUZ
 
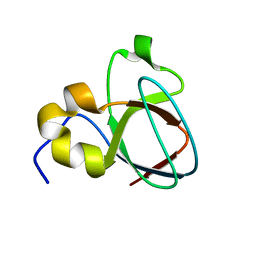 | |
1MV0
 
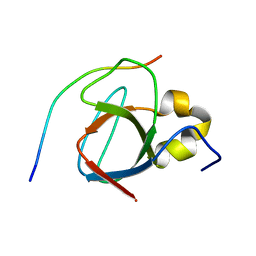 | |
1JRM
 
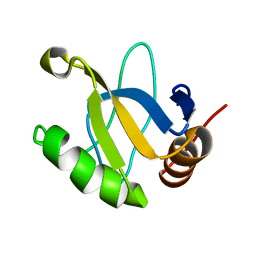 | |
1X9A
 
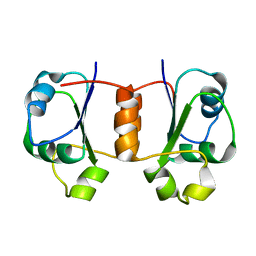 | | Solution NMR Structure of Protein Tm0979 from Thermotoga maritima. Ontario Center for Structural Proteomics Target TM0979_1_87; Northeast Structural Genomics Consortium Target VT98. | | Descriptor: | hypothetical protein TM0979 | | Authors: | Gaspar, J.A, Liu, C, Vassall, K.A, Stathopulos, P.B, Meglei, G, Stephen, R, Pineda-Lucena, A, Wu, B, Yee, A, Arrowsmith, C.H, Meiering, E.M, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-08-20 | | Release date: | 2004-12-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A novel member of the YchN-like fold: solution structure of the hypothetical protein Tm0979 from Thermotoga maritima.
Protein Sci., 14, 2005
|
|
2L6J
 
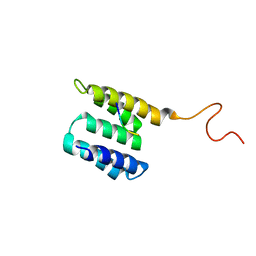 | | Tah1 complexed by MEEVD | | Descriptor: | C-terminus Hsp90 chaperone peptide MEEVD, TPR repeat-containing protein associated with Hsp90 | | Authors: | Jimenez, B, Ugwu, F, Zhao, R, Orti, L, Houry, W.A, Pineda-Lucena, A. | | Deposit date: | 2010-11-22 | | Release date: | 2011-12-28 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of minimal tetratricopeptide repeat domain protein Tah1 reveals mechanism of its interaction with Pih1 and Hsp90.
J.Biol.Chem., 287, 2012
|
|
1RYJ
 
 | | Solution NMR Structure of Protein Mth1743 from Methanobacterium thermoautotrophicum. Ontario Centre for Structural Proteomics target MTH1743_1_70; Northeast Structural Genomics Consortium Target TT526. | | Descriptor: | unknown | | Authors: | Yee, A, Chang, X, Pineda-Lucena, A, Wu, B, Semesi, A, Le, B, Ramelot, T, Lee, G.M, Bhattacharyya, S, Gutierrez, P, Denisov, A, Lee, C.H, Cort, J.R, Kozlov, G, Liao, J, Finak, G, Chen, L, Wishart, D, Lee, W, McIntosh, L.P, Gehring, K, Kennedy, M.A, Edwards, A.M, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-12-22 | | Release date: | 2004-02-24 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | AN NMR APPROACH TO STRUCTURAL PROTEOMICS
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
2B3G
 
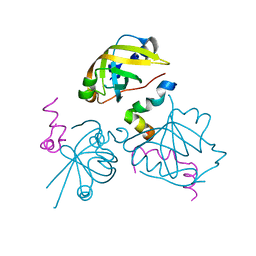 | | p53N (fragment 33-60) bound to RPA70N | | Descriptor: | Cellular tumor antigen p53, Replication protein A 70 kDa DNA-binding subunit | | Authors: | Bochkareva, E, Kaustov, L, Ayed, A, Yi, G.S, Lu, Y, Pineda-Lucena, A, Liao, J.C, Okorokov, A.L, Milner, J, Arrowsmith, C.H, Bochkarev, A. | | Deposit date: | 2005-09-20 | | Release date: | 2005-10-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Single-stranded DNA mimicry in the p53 transactivation domain interaction with replication protein A.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1DZC
 
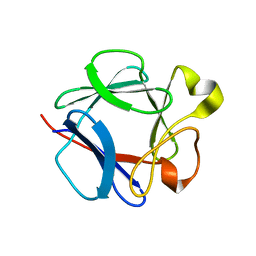 | | High resolution structure of acidic fibroblast growth factor. Mutant FGF-4-ALA-(24-154), 24 NMR structures | | Descriptor: | FIBROBLAST GROWTH FACTOR 1 | | Authors: | Lozano, R.M, Pineda-Lucena, A, Gonzalez, C, Jimenez, M.A, Cuevas, P, Redondo-Horcajo, M, Sanz, J.M, Rico, M, Gimenez-Gallego, G. | | Deposit date: | 2000-02-24 | | Release date: | 2000-03-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | 1H-NMR Structural Characterization of a Non Mitogenic, Vasodilatory, Ischemia-Protector and Neuromodulatory Acidic Fibroblast Growth Factor
Biochemistry, 39, 2000
|
|
1DZD
 
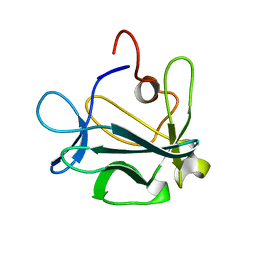 | | High resolution structure of acidic fibroblast growth factor (27-154), 24 NMR structures | | Descriptor: | ACIDIC FIBROBLAST GROWTH FACTOR | | Authors: | Lozano, R.M, Pineda-Lucena, A, Gonzalez, C, Jimenez, M.A, Cuevas, P, Redondo-Horcajo, M, Sanz, J.M, Rico, M, Gimenez-Gallego, G. | | Deposit date: | 2000-02-24 | | Release date: | 2000-03-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | 1H-NMR Structural Characterization of a Non Mitogenic, Vasodilatory, Ischemia-Protector and Neuromodulatory Acidic Fibroblast Growth Factor
Biochemistry, 39, 2000
|
|
2W9O
 
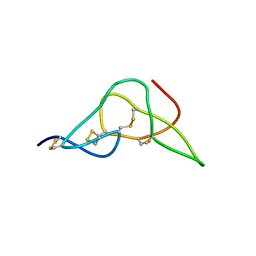 | | Solution structure of jerdostatin from Trimeresurus jerdonii | | Descriptor: | SHORT DISINTEGRIN JERDOSTATIN | | Authors: | Carbajo, R.J, Sanz, L, Mosulen, S, Calvete, J.J, Pineda-Lucena, A. | | Deposit date: | 2009-01-27 | | Release date: | 2010-03-31 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | NMR Structure and Dynamics of Recombinant Wild-Type and Mutated Jerdostatin, a Selective Inhibitor of Integrin Alpha1 Beta1
Proteins, 79, 2011
|
|
2W9W
 
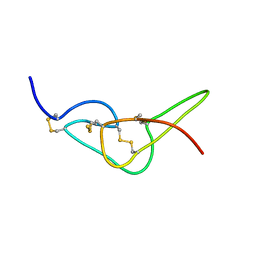 | | Solution structure of jerdostatin mutant R24K from Trimeresurus jerdonii with end C-terminal residues N45G46 deleted | | Descriptor: | SHORT DISINTEGRIN JERDOSTATIN | | Authors: | Carbajo, R.J, Sanz, L, Mosulen, S, Calvete, J.J, Pineda-Lucena, A. | | Deposit date: | 2009-01-29 | | Release date: | 2010-03-31 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | NMR Structure and Dynamics of Recombinant Wild-Type and Mutated Jerdostatin, a Selective Inhibitor of Integrin Alpha1 Beta1
Proteins, 79, 2011
|
|
2W9V
 
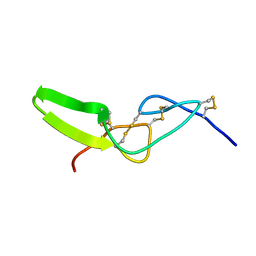 | | Solution structure of jerdostatin from Trimeresurus jerdonii with end C-terminal residues N45G46 deleted | | Descriptor: | SHORT DISINTEGRIN JERDOSTATIN | | Authors: | Carbajo, R.J, Sanz, L, Mosulen, S, Calvete, J.J, Pineda-Lucena, A. | | Deposit date: | 2009-01-29 | | Release date: | 2010-03-31 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | NMR Structure and Dynamics of Recombinant Wild-Type and Mutated Jerdostatin, a Selective Inhibitor of Integrin Alpha1 Beta1
Proteins, 79, 2011
|
|
2W9U
 
 | | Solution structure of jerdostatin mutant R24K from Trimeresurus jerdonii | | Descriptor: | SHORT DISINTEGRIN JERDOSTATIN | | Authors: | Carbajo, R.J, Sanz, L, Mosulen, S, Calvete, J.J, Pineda-Lucena, A. | | Deposit date: | 2009-01-29 | | Release date: | 2010-03-31 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | NMR Structure and Dynamics of Recombinant Wild-Type and Mutated Jerdostatin, a Selective Inhibitor of Integrin Alpha1 Beta1
Proteins, 79, 2011
|
|
1NE3
 
 | | Solution structure of ribosomal protein S28E from Methanobacterium Thermoautotrophicum. Ontario Centre for Structural Proteomics target MTH0256_1_68; Northeast Structural Genomics Target TT744 | | Descriptor: | 30S ribosomal protein S28E | | Authors: | Wu, B, Pineda-Lucena, A, Yee, A, Cort, J.R, Ramelot, T.A, Kennedy, M, Edwards, A, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2002-12-10 | | Release date: | 2003-12-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of ribosomal protein S28E from Methanobacterium thermoautotrophicum.
Protein Sci., 12, 2003
|
|
1NEI
 
 | | Solution NMR Structure of Protein yoaG from Escherichia coli. Ontario Centre for Structural Proteomics Target EC0264_1_60; Northeast Structural Genomics Consortium Target ET94. | | Descriptor: | hypothetical protein yoaG | | Authors: | Wu, B, Pineda-Lucena, A, Yee, A, Cort, J, Kennedy, M.A, Edwards, A.M, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2002-12-11 | | Release date: | 2004-04-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of hypothetical protein dimer encoded by the Yoag gene from Escherichia coli
To be published
|
|
2I9N
 
 | |
2J10
 
 | | p53 tetramerization domain mutant T329F Q331K | | Descriptor: | CELLULAR TUMOR ANTIGEN P53 | | Authors: | Carbajo, R.J, Mora, P, Sanchez del Pino, M.M, Perez-Paya, E, Pineda-Lucena, A. | | Deposit date: | 2006-08-08 | | Release date: | 2007-08-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solvent-exposed residues located in the beta-sheet modulate the stability of the tetramerization domain of p53--a structural and combinatorial approach.
Proteins, 71, 2008
|
|
2J0Z
 
 | | p53 tetramerization domain wild type | | Descriptor: | CELLULAR TUMOR ANTIGEN P53 | | Authors: | Carbajo, R.J, Mora, P, Sanchez del Pino, M.M, Perez-Paya, E, Pineda-Lucena, A. | | Deposit date: | 2006-08-08 | | Release date: | 2007-08-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solvent-exposed residues located in the beta-sheet modulate the stability of the tetramerization domain of p53--a structural and combinatorial approach.
Proteins, 71, 2008
|
|
2I9M
 
 | |
2J11
 
 | | p53 tetramerization domain mutant Y327S T329G Q331G | | Descriptor: | CELLULAR TUMOR ANTIGEN P53 | | Authors: | Carbajo, R.J, Mora, P, Sanchez del Pino, M.M, Perez-Paya, E, Pineda-Lucena, A. | | Deposit date: | 2006-08-08 | | Release date: | 2007-08-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solvent-exposed residues located in the beta-sheet modulate the stability of the tetramerization domain of p53--a structural and combinatorial approach.
Proteins, 71, 2008
|
|
1MP1
 
 | | Solution structure of the PWI motif from SRm160 | | Descriptor: | Ser/Arg-related nuclear matrix protein | | Authors: | Szymczyna, B.R, Bowman, J, McCracken, S, Pineda-Lucena, A, Lu, Y, Cox, B, Lambermon, M, Graveley, B.R, Arrowsmith, C.H, Blencowe, B.J. | | Deposit date: | 2002-09-11 | | Release date: | 2003-09-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and function of the PWI motif: a novel nucleic acid-binding domain that facilitates pre-mRNA processing.
Genes Dev., 17, 2003
|
|
1RML
 
 | | NMR STUDY OF ACID FIBROBLAST GROWTH FACTOR BOUND TO 1,3,6-NAPHTHALENE TRISULPHONATE, 26 STRUCTURES | | Descriptor: | ACIDIC FIBROBLAST GROWTH FACTOR, NAPHTHALENE TRISULFONATE | | Authors: | Lozano, R.M, Jimenez, M.A, Santoro, J, Rico, M, Gimenez-Gallego, G. | | Deposit date: | 1998-05-21 | | Release date: | 1998-11-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of acidic fibroblast growth factor bound to 1,3, 6-naphthalenetrisulfonate: a minimal model for the anti-tumoral action of suramins and suradistas.
J.Mol.Biol., 281, 1998
|
|
