1EU5
 
 | | STRUCTURE OF E. COLI DUTPASE AT 1.45 A | | Descriptor: | DEOXYURIDINE 5'-TRIPHOSPHATE NUCLEOTIDOHYDROLASE, GLYCEROL | | Authors: | Gonzalez, A, Larsson, G, Persson, R, Cedergren-Zeppezauer, E. | | Deposit date: | 2000-04-13 | | Release date: | 2000-05-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Atomic resolution structure of Escherichia coli dUTPase determined ab initio.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1DUN
 
 | | EIAV DUTPASE NATIVE | | Descriptor: | DEOXYURIDINE 5'-TRIPHOSPHATE NUCLEOTIDOHYDROLASE | | Authors: | Dauter, Z, Persson, R, Rosengren, A.M, Nyman, P.O, Wilson, K.S, Cedergren-Zeppezauer, E.S. | | Deposit date: | 1997-11-27 | | Release date: | 1998-05-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of dUTPase from equine infectious anaemia virus; active site metal binding in a substrate analogue complex.
J.Mol.Biol., 285, 1999
|
|
1DUC
 
 | | EIAV DUTPASE DUDP/STRONTIUM COMPLEX | | Descriptor: | DEOXYURIDINE 5'-TRIPHOSPHATE NUCLEOTIDOHYDROLASE, DEOXYURIDINE-5'-DIPHOSPHATE, STRONTIUM ION | | Authors: | Dauter, Z, Persson, R, Rosengren, A.M, Nyman, P.O, Wilson, K.S, Cedergren-Zeppezauer, E.S. | | Deposit date: | 1997-11-29 | | Release date: | 1998-06-03 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of dUTPase from equine infectious anaemia virus; active site metal binding in a substrate analogue complex.
J.Mol.Biol., 285, 1999
|
|
1EUW
 
 | | ATOMIC RESOLUTION STRUCTURE OF E. COLI DUTPASE | | Descriptor: | DEOXYURIDINE 5'-TRIPHOSPHATE NUCLEOTIDOHYDROLASE, ETHYL MERCURY ION, GLYCEROL | | Authors: | Gonzalez, A, Cedergren, E, Larsson, G, Persson, R. | | Deposit date: | 2000-04-17 | | Release date: | 2000-05-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Atomic resolution structure of Escherichia coli dUTPase determined ab initio.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
2XX6
 
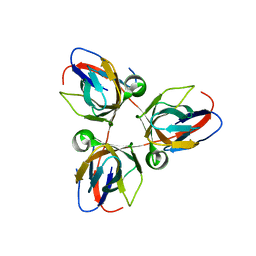 | | Structure of the Bacillus subtilis prophage dUTPase, YosS | | Descriptor: | SPBC2 PROPHAGE-DERIVED DEOXYURIDINE 5'-TRIPHOSPHATE NUCLEOTIDOHYDROLASE YOSS | | Authors: | Garcia-Nafria, J, Harkiolaki, M, Persson, R, Fogg, M.J, Wilson, K.S. | | Deposit date: | 2010-11-08 | | Release date: | 2011-02-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | The Structure of Bacillus Subtilis Sp Beta Prophage Dutpase and its Complexes with Two Nucleotides
Acta Crystallogr.,Sect.D, 67, 2011
|
|
2Y1T
 
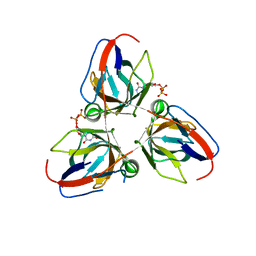 | | Bacillus subtilis prophage dUTPase in complex with dUDP | | Descriptor: | DEOXYURIDINE-5'-DIPHOSPHATE, SPBC2 PROPHAGE-DERIVED DEOXYURIDINE 5'-TRIPHOSPHATE NUCLEOTIDOHYDROLASE YOSS | | Authors: | Garcia-Nafria, J, Harkiolaki, M, Persson, R, Fogg, M.J, Wilson, K.S. | | Deposit date: | 2010-12-10 | | Release date: | 2011-02-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | The Structure of Bacillus Subtilis Sp Beta Prophage Dutpase and its Complexes with Two Nucleotides
Acta Crystallogr.,Sect.D, 67, 2011
|
|
2XY3
 
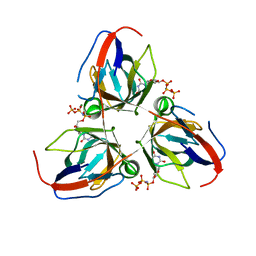 | | Structure of the Bacillus subtilis prophage dUTPase with dUpNHpp | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, MAGNESIUM ION, SPBC2 PROPHAGE-DERIVED DEOXYURIDINE 5'-TRIPHOSPHATE NUCLEOTIDOHYDROLASE YOSS | | Authors: | Garcia-Nafria, J, Harkiolaki, M, Persson, R, Fogg, M.J, Wilson, K.S. | | Deposit date: | 2010-11-12 | | Release date: | 2011-02-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | The Structure of Bacillus Subtilis Sp Beta Prophage Dutpase and its Complexes with Two Nucleotides
Acta Crystallogr.,Sect.D, 67, 2011
|
|
1SYL
 
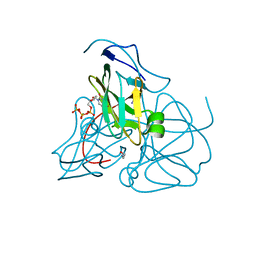 | | Crystal structure of inactive mutant dUTPase complexed with substrate dUTP | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DEOXYURIDINE-5'-TRIPHOSPHATE, Deoxyuridine 5'-triphosphate nucleotidohydrolase, ... | | Authors: | Barabas, O, Kovari, J, Pongracz, V, Wilmanns, M, Vertessy, B.G. | | Deposit date: | 2004-04-01 | | Release date: | 2004-09-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Insights into the Catalytic Mechanism of Phosphate Ester Hydrolysis by dUTPase
J.Biol.Chem., 279, 2004
|
|
1RNJ
 
 | | Crystal structure of inactive mutant dUTPase complexed with substrate analogue imido-dUTP | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Deoxyuridine 5'-triphosphate nucleotidohydrolase, ... | | Authors: | Barabas, O, Pongracz, V, Kovari, J, Wilmanns, M, Vertessy, B.G. | | Deposit date: | 2003-12-01 | | Release date: | 2004-09-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Insights into the Catalytic Mechanism of Phosphate Ester Hydrolysis by dUTPase.
J.Biol.Chem., 279, 2004
|
|
1RN8
 
 | | Crystal structure of dUTPase complexed with substrate analogue imido-dUTP | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, ... | | Authors: | Barabas, O, Pongracz, V, Kovari, J, Wilmanns, M, Vertessy, B.G. | | Deposit date: | 2003-12-01 | | Release date: | 2004-09-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural Insights into the Catalytic Mechanism of Phosphate Ester Hydrolysis by dUTPase.
J.Biol.Chem., 279, 2004
|
|
1SEH
 
 | | Crystal structure of E. coli dUTPase complexed with the product dUMP | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Deoxyuridine 5'-triphosphate nucleotidohydrolase | | Authors: | Barabas, O, Kovari, J, Pongracz, V, Wilmanns, M, Vertessy, B.G. | | Deposit date: | 2004-02-17 | | Release date: | 2004-09-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structural Insights into the Catalytic Mechanism of Phosphate Ester Hydrolysis by dUTPase
J.Biol.Chem., 279, 2004
|
|
2HR6
 
 | | Crystal structure of dUTPase in complex with substrate analogue dUDP and manganese | | Descriptor: | 1,2-ETHANEDIOL, DEOXYURIDINE-5'-DIPHOSPHATE, Deoxyuridine 5'-triphosphate nucleotidohydrolase, ... | | Authors: | Barabas, O, Kovari, J, Tapai, R, Vertessy, B.G. | | Deposit date: | 2006-07-19 | | Release date: | 2007-07-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Methylene substitution at the alpha-beta bridging position within the phosphate chain of dUDP profoundly perturbs ligand accommodation into the dUTPase active site.
Proteins, 71, 2008
|
|
2HRM
 
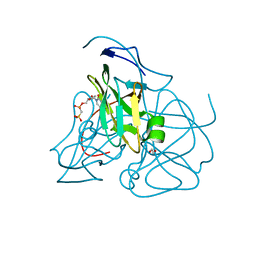 | | Crystal structure of dUTPase complexed with substrate analogue methylene-dUTP | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXY-5'-O-[(S)-HYDROXY(PHOSPHONOMETHYL)PHOSPHORYL]URIDINE, Deoxyuridine 5'-triphosphate nucleotidohydrolase | | Authors: | Barabas, O, Kovari, J, Tapai, R, Vertessy, B.G. | | Deposit date: | 2006-07-20 | | Release date: | 2007-07-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Methylene substitution at the alpha-beta bridging position within the phosphate chain of dUDP profoundly perturbs ligand accommodation into the dUTPase active site.
Proteins, 71, 2008
|
|
