1CTN
 
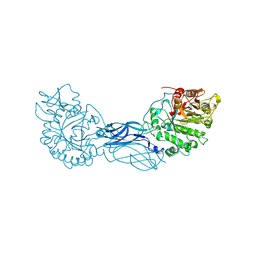 | | CRYSTAL STRUCTURE OF A BACTERIAL CHITINASE AT 2.3 ANGSTROMS RESOLUTION | | Descriptor: | CHITINASE A | | Authors: | Perrakis, A, Tews, I, Dauter, Z, Wilson, K.S, Vorgias, C.E. | | Deposit date: | 1994-10-10 | | Release date: | 1995-02-07 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a bacterial chitinase at 2.3 A resolution.
Structure, 2, 1994
|
|
6QCG
 
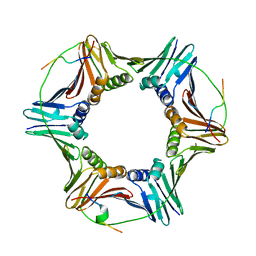 | |
8ARF
 
 | |
2KLS
 
 | | Apo-form of the second Ca2+ binding domain of NCX1.4 | | Descriptor: | Sodium/calcium exchanger 1 | | Authors: | Hilge, M, Aelen, J, Foarce, A, Perrakis, A, Vuister, G.W. | | Deposit date: | 2009-07-08 | | Release date: | 2009-08-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Ca2+ regulation in the Na+/Ca2+ exchanger features a dual electrostatic switch mechanism.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3KJY
 
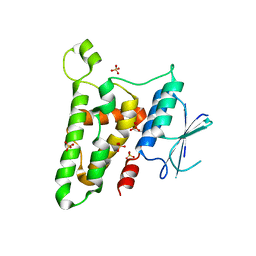 | | Crystal structure of reduced HOMO SAPIENS CLIC3 | | Descriptor: | Chloride intracellular channel protein 3, SULFATE ION | | Authors: | Littler, D.R, Curmi, P.M.G, Breit, S.N, Perrakis, A. | | Deposit date: | 2009-11-04 | | Release date: | 2009-11-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of human CLIC3 at 2 A resolution
Proteins, 78, 2010
|
|
7P0K
 
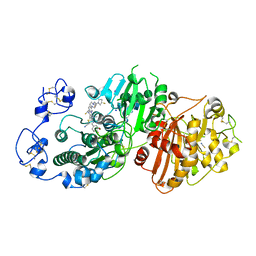 | | Crystal structure of Autotaxin (ENPP2) with 18F-labeled positron emission tomography ligand | | Descriptor: | 2-[[2-ethyl-6-[4-[2-[(3~{R})-3-fluoranylpyrrolidin-1-yl]-2-oxidanylidene-ethyl]piperazin-1-yl]imidazo[1,2-a]pyridin-3-yl]-methyl-amino]-4-(4-fluorophenyl)-2,3-dihydro-1,3-thiazole-5-carbonitrile, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Salgado-Polo, F, Shao, T, Xiao, Z, Van, R, Chen, J, Rong, J, Haider, A, Shao, Y, Josephson, L, Perrakis, A, Liang, S.H. | | Deposit date: | 2021-06-29 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Imaging Autotaxin In Vivo with 18 F-Labeled Positron Emission Tomography Ligands
J Med Chem, 64, 2021
|
|
6NVQ
 
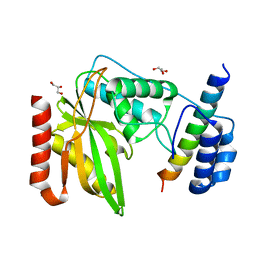 | |
2XR9
 
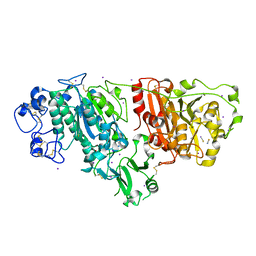 | | Crystal structure of Autotaxin (ENPP2) | | Descriptor: | CALCIUM ION, ECTONUCLEOTIDE PYROPHOSPHATASE/PHOSPHODIESTERASE FAMILY MEMBER 2, IODIDE ION, ... | | Authors: | Kamtekar, S, Hausmann, J, Day, J.E, Christodoulou, E, Perrakis, A. | | Deposit date: | 2010-09-13 | | Release date: | 2011-01-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis of substrate discrimination and integrin binding by autotaxin.
Nat. Struct. Mol. Biol., 18, 2011
|
|
1QBB
 
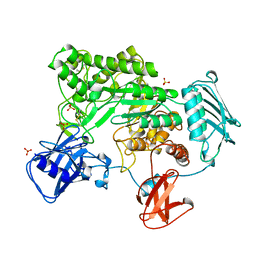 | | BACTERIAL CHITOBIASE COMPLEXED WITH CHITOBIOSE (DINAG) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHITOBIASE, SULFATE ION | | Authors: | Tews, I, Perrakis, A, Oppenheim, A, Dauter, Z, Wilson, K.S, Vorgias, C.E. | | Deposit date: | 1996-06-07 | | Release date: | 1997-02-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Bacterial chitobiase structure provides insight into catalytic mechanism and the basis of Tay-Sachs disease.
Nat.Struct.Biol., 3, 1996
|
|
1QBA
 
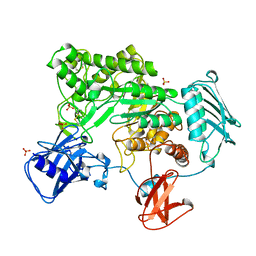 | | BACTERIAL CHITOBIASE, GLYCOSYL HYDROLASE FAMILY 20 | | Descriptor: | CHITOBIASE, SULFATE ION | | Authors: | Tews, I, Perrakis, A, Oppenheim, A, Dauter, Z, Wilson, K.S, Vorgias, C.E. | | Deposit date: | 1996-06-06 | | Release date: | 1997-01-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Bacterial chitobiase structure provides insight into catalytic mechanism and the basis of Tay-Sachs disease.
Nat.Struct.Biol., 3, 1996
|
|
3G73
 
 | | Structure of the FOXM1 DNA binding | | Descriptor: | DNA (5'-D(P*AP*AP*AP*TP*TP*GP*TP*TP*TP*AP*TP*AP*AP*AP*CP*AP*GP*CP*CP*CP*G)-3'), DNA (5'-D(P*TP*TP*CP*GP*GP*GP*CP*TP*GP*TP*TP*TP*AP*TP*AP*AP*AP*CP*AP*AP*T)-3'), Forkhead box protein M1, ... | | Authors: | Littler, D.R, Perrakis, A, Hibbert, R.G, Medema, R.H. | | Deposit date: | 2009-02-09 | | Release date: | 2009-03-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structure of the FoxM1 DNA-recognition domain bound to a promoter sequence
Nucleic Acids Res., 2010
|
|
2WVR
 
 | | Human Cdt1:Geminin complex | | Descriptor: | DNA REPLICATION FACTOR CDT1, GEMININ | | Authors: | De Marco, V, Perrakis, A. | | Deposit date: | 2009-10-19 | | Release date: | 2009-10-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Quaternary Structure of the Human Cdt1-Geminin Complex Regulates DNA Replication Licensing.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
7Z5G
 
 | | human apo MATCAP | | Descriptor: | Uncharacterized protein KIAA0895-like | | Authors: | Bak, J, Adamoupolos, A, Heidebrecht, T, Perrakis, A. | | Deposit date: | 2022-03-09 | | Release date: | 2022-05-11 | | Last modified: | 2022-06-01 | | Method: | X-RAY DIFFRACTION (2.113 Å) | | Cite: | Posttranslational modification of microtubules by the MATCAP detyrosinase.
Science, 376, 2022
|
|
7Z5H
 
 | | human Zn MATCAP | | Descriptor: | Uncharacterized protein KIAA0895-like, ZINC ION | | Authors: | Bak, J, Adamopoulos, A, Heidebrecht, T, Perrakis, A. | | Deposit date: | 2022-03-09 | | Release date: | 2022-05-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Posttranslational modification of microtubules by the MATCAP detyrosinase.
Science, 376, 2022
|
|
7Z6S
 
 | | MATCAP bound to a human 14 protofilament microtubule | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Bak, J, Perrakis, A. | | Deposit date: | 2022-03-14 | | Release date: | 2022-05-11 | | Last modified: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Posttranslational modification of microtubules by the MATCAP detyrosinase.
Science, 376, 2022
|
|
1VYB
 
 | | Endonuclease domain of human LINE1 ORF2p | | Descriptor: | GLYCEROL, ORF2 CONTAINS A REVERSE TRANSCRIPTASE DOMAIN, SULFATE ION, ... | | Authors: | Weichenrieder, O, Repanas, K, Perrakis, A. | | Deposit date: | 2004-04-25 | | Release date: | 2004-06-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the targeting endonuclease of the human LINE-1 retrotransposon.
Structure, 12, 2004
|
|
7Z0N
 
 | | Structure-Based Design of a Novel Class of Autotaxin Inhibitors Based on Endogenous Allosteric Modulators | | Descriptor: | CALCIUM ION, GLYCEROL, IODIDE ION, ... | | Authors: | Salgado-Polo, F, Clark, J.M, Macdonald, S.J.F, Barrett, T.N, Perrakis, A, Jamieson, A. | | Deposit date: | 2022-02-23 | | Release date: | 2022-05-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-Based Design of a Novel Class of Autotaxin Inhibitors Based on Endogenous Allosteric Modulators.
J.Med.Chem., 65, 2022
|
|
2CKL
 
 | | Ring1b-Bmi1 E3 catalytic domain structure | | Descriptor: | IODIDE ION, POLYCOMB GROUP RING FINGER PROTEIN 4, UBIQUITIN LIGASE PROTEIN RING2, ... | | Authors: | Buchwald, G, van der Stoop, P, Weichenrieder, O, Perrakis, A, van Lohuizen, M, Sixma, T.K. | | Deposit date: | 2006-04-20 | | Release date: | 2006-05-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and E3-Ligase Activity of the Ring-Ring Complex of Polycomb Proteins Bmi1 and Ring1B.
Embo J., 25, 2006
|
|
3FY7
 
 | | Crystal structure of homo sapiens CLIC3 | | Descriptor: | Chloride intracellular channel protein 3, SULFATE ION | | Authors: | Littler, D.R, Curmi, P.M.G, Breit, S.N, Perrakis, A. | | Deposit date: | 2009-01-22 | | Release date: | 2009-02-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of human CLIC3 at 2 A resolution
Proteins, 78, 2010
|
|
6FPV
 
 | | A llama-derived JBP1-targeting nanobody | | Descriptor: | GLYCEROL, Nanobody | | Authors: | van Beusekom, B, Adamopoulos, A, Heidebrecht, T, Joosten, R.P, Perrakis, A. | | Deposit date: | 2018-02-12 | | Release date: | 2018-10-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Characterization and structure determination of a llama-derived nanobody targeting the J-base binding protein 1.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
2X4S
 
 | | Crystal structure of MHC CLass I HLA-A2.1 bound to a peptide representing the epitope of the H5N1 (Avian Flu) Nucleoprotein | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, BETA-2-MICROGLOBULIN, GLYCEROL, ... | | Authors: | Celie, P.H.N, Toebes, M, Rodenko, B, Ovaa, H, Perrakis, A, Schumacher, T.N.M. | | Deposit date: | 2010-02-02 | | Release date: | 2010-03-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Uv-Induced Ligand Exchange in Mhc Class I Protein Crystals
J.Am.Chem.Soc., 131, 2009
|
|
2X4O
 
 | | Crystal structure of MHC CLass I HLA-A2.1 bound to HIV-1 envelope peptide env120-128 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, BETA-2-MICROGLOBULIN, ENVELOPE GLYCOPROTEIN GP160, ... | | Authors: | Celie, P.H.N, Toebes, M, Rodenko, B, Ovaa, H, Perrakis, A, Schumacher, T.N.M. | | Deposit date: | 2010-02-02 | | Release date: | 2010-03-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Uv-Induced Ligand Exchange in Mhc Class I Protein Crystals.
J.Am.Chem.Soc., 131, 2009
|
|
2XSE
 
 | | The structural basis for recognition of J-base containing DNA by a novel DNA-binding domain in JBP1 | | Descriptor: | GLYCEROL, NITRATE ION, THYMINE DIOXYGENASE JBP1 | | Authors: | Heidebrecht, T, Christodoulou, E, Chalmers, M.J, Jan, S, ter Riete, B, Grover, R.K, Joosten, R.P, Littler, D, vanLuenen, H, Griffin, P.R, Wentworth, P, Borst, P, Perrakis, A. | | Deposit date: | 2010-09-28 | | Release date: | 2011-03-30 | | Last modified: | 2011-08-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Structural Basis for Recognition of Base J Containing DNA by a Novel DNA Binding Domain in Jbp1.
Nucleic Acids Res., 39, 2011
|
|
2XRG
 
 | | Crystal structure of Autotaxin (ENPP2) in complex with the HA155 boronic acid inhibitor | | Descriptor: | CALCIUM ION, ECTONUCLEOTIDE PYROPHOSPHATASE/PHOSPHODIESTERASE FAMILY MEMBER 2, IODIDE ION, ... | | Authors: | Hausmann, J, Albers, H.M.H.G, Perrakis, A. | | Deposit date: | 2010-09-14 | | Release date: | 2011-01-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural Basis of Substrate Discrimination and Integrin Binding by Autotaxin.
Nat.Struct.Mol.Biol., 18, 2011
|
|
2Y7Y
 
 | | APLYSIA CALIFORNICA ACHBP IN APO STATE | | Descriptor: | SOLUBLE ACETYLCHOLINE RECEPTOR | | Authors: | Ulens, C, Akdemir, A, Jongejan, A, van Elk, R, Bertrand, S, Perrakis, A, Leurs, R, Smit, A.B, Sixma, T.K, Bertrand, D, De Esch, I.J. | | Deposit date: | 2011-02-02 | | Release date: | 2011-03-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.895 Å) | | Cite: | Use of Acetylcholine Binding Protein in the Search for Novel Alpha7 Nicotinic Receptor Ligands. In Silico Docking, Pharmacological Screening, and X-Ray Analysis.
J.Med.Chem., 52, 2009
|
|
