7SHF
 
 | | Cryo-EM structure of GPR158 coupled to the RGS7-Gbeta5 complex | | Descriptor: | (2S)-1-{[(S)-hydroxy{[(1s,2R,3R,4R,5S,6S)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}-3-(octadecanoyloxy)propan-2-yl (5E,8E,11E,14E)-icosa-5,8,11,14-tetraenoate, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, CHOLESTEROL, ... | | Authors: | Patil, D.N, Singh, S, Singh, A.K, Martemyanov, K.A. | | Deposit date: | 2021-10-08 | | Release date: | 2021-12-01 | | Last modified: | 2022-01-19 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structure of human GPR158 receptor coupled to the RGS7-G beta 5 signaling complex.
Science, 375, 2022
|
|
7SHE
 
 | | Cryo-EM structure of human GPR158 | | Descriptor: | (2S)-1-{[(S)-hydroxy{[(1s,2R,3R,4R,5S,6S)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}-3-(octadecanoyloxy)propan-2-yl (5E,8E,11E,14E)-icosa-5,8,11,14-tetraenoate, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, CHOLESTEROL, ... | | Authors: | Patil, D.N, Singh, S, Singh, A.K, Martemyanov, K.A. | | Deposit date: | 2021-10-08 | | Release date: | 2021-12-01 | | Last modified: | 2022-01-19 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structure of human GPR158 receptor coupled to the RGS7-G beta 5 signaling complex.
Science, 375, 2022
|
|
6N9G
 
 | | Crystal Structure of RGS7-Gbeta5 dimer | | Descriptor: | Guanine nucleotide-binding protein subunit beta-5, Regulator of G-protein signaling 7 | | Authors: | Patil, D.N, Rangarajan, E, Izard, T, Martemyanov, K.A. | | Deposit date: | 2018-12-03 | | Release date: | 2019-01-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.129 Å) | | Cite: | Structural organization of a major neuronal G protein regulator, the RGS7-G beta 5-R7BP complex.
Elife, 7, 2018
|
|
8D1B
 
 | |
4AN6
 
 | |
4AN7
 
 | |
4B15
 
 | | crystal structure of tamarind chitinase like lectin (TCLL) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Patil, D.N, Kumar, P. | | Deposit date: | 2012-07-06 | | Release date: | 2013-06-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structural Investigation of a Novel N-Acetyl Glucosamine Binding Chi-Lectin which Reveals Evolutionary Relationship with Class III Chitinases.
Plos One, 8, 2013
|
|
4B16
 
 | |
3ZQ7
 
 | |
2Y99
 
 | | Crystal Structure of cis-Biphenyl-2,3-dihydrodiol-2,3-dehydrogenase (BphB)from Pandoraea pnomenusa strain B-356 complex with co-enzyme NAD | | Descriptor: | CIS-2,3-DIHYDROBIPHENYL-2,3-DIOL DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Dhindwal, S, Patil, D.N, Kumar, P. | | Deposit date: | 2011-02-12 | | Release date: | 2011-08-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Biochemical Studies and Ligand-Bound Structures of Biphenyl Dehydrogenase from Pandoraea Pnomenusa Strain B-356 Reveal a Basis for Broad Specificity of the Enzyme.
J.Biol.Chem., 286, 2011
|
|
2Y93
 
 | | Crystal Structure of cis-Biphenyl-2,3-dihydrodiol-2,3-dehydrogenase (BphB)from Pandoraea pnomenusa strain B-356. | | Descriptor: | CIS-2,3-DIHYDROBIPHENYL-2,3-DIOL DEHYDROGENASE | | Authors: | Dhindwal, S, Patil, D.N, Kumar, P. | | Deposit date: | 2011-02-11 | | Release date: | 2011-08-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Biochemical Studies and Ligand-Bound Structures of Biphenyl Dehydrogenase from Pandoraea Pnomenusa Strain B-356 Reveal a Basis for Broad Specificity of the Enzyme.
J.Biol.Chem., 286, 2011
|
|
3ZV3
 
 | | CRYSTAL STRUCTURE OF CIS-BIPHENYL-2,3-DIHYDRODIOL-2,3-DEHYDROGENASE (BPHB)FROM PANDORAEA PNOMENUSA STRAIN B-356 IN INTERMEDIATE STATE OF SUBSTRATE BINDING LOOP | | Descriptor: | CIS-2,3-DIHYDROBIPHENYL-2,3-DIOL DEHYDROGENASE | | Authors: | Dhindwal, S, Patil, D.N, Kumar, P. | | Deposit date: | 2011-07-23 | | Release date: | 2011-08-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Biochemical Studies and Ligand-Bound Structures of Biphenyl Dehydrogenase from Pandoraea Pnomenusa Strain B-356 Reveal a Basis for Broad Specificity of the Enzyme.
J.Biol.Chem., 286, 2011
|
|
3ZV6
 
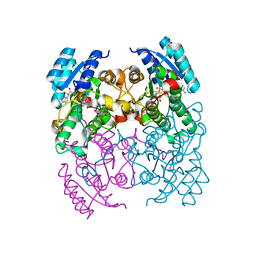 | | CRYSTAL STRUCTURE OF CIS-BIPHENYL-2,3-DIHYDRODIOL-2,3-DEHYDROGENASE (BPHB) FROM PANDORAEA PNOMENUSA STRAIN B-356 COMPLEX WITH CO-ENZYME NAD AND PRODUCT ANALOG 4,4'-DIHYDROXYBIPHENYL | | Descriptor: | 4,4'-DIHYDROXYBIPHENYL, CIS-2,3-DIHYDROBIPHENYL-2,3-DIOL DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Dhindwal, S, Patil, D.N, Kumar, P. | | Deposit date: | 2011-07-23 | | Release date: | 2011-08-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Biochemical Studies and Ligand-Bound Structures of Biphenyl Dehydrogenase from Pandoraea Pnomenusa Strain B-356 Reveal a Basis for Broad Specificity of the Enzyme.
J.Biol.Chem., 286, 2011
|
|
3ZV5
 
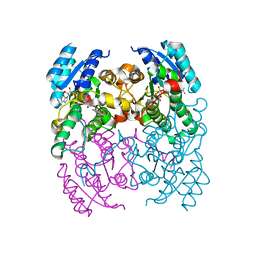 | | CRYSTAL STRUCTURE OF CIS-BIPHENYL-2,3-DIHYDRODIOL-2,3-DEHYDROGENASE (BPHB) FROM PANDORAEA PNOMENUSA STRAIN B-356 COMPLEX WITH CO-ENZYME NAD AND PRODUCT 2,3-DIHYDROXYBIPHENYL | | Descriptor: | BIPHENYL-2,3-DIOL, CIS-2,3-DIHYDROBIPHENYL-2,3-DIOL DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Dhindwal, S, Patil, D.N, Kumar, P. | | Deposit date: | 2011-07-23 | | Release date: | 2011-08-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Biochemical Studies and Ligand-Bound Structures of Biphenyl Dehydrogenase from Pandoraea Pnomenusa Strain B-356 Reveal a Basis for Broad Specificity of the Enzyme.
J.Biol.Chem., 286, 2011
|
|
3ZV4
 
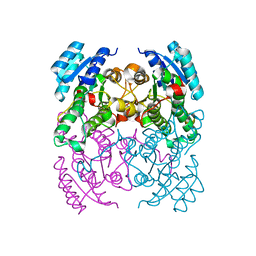 | | CRYSTAL STRUCTURE OF CIS-BIPHENYL-2,3-DIHYDRODIOL-2,3-DEHYDROGENASE (BPHB) FROM PANDORAEA PNOMENUSA STRAIN B-356 IN APO FORM AT 1.8 ANGSTROM | | Descriptor: | CIS-2,3-DIHYDROBIPHENYL-2,3-DIOL DEHYDROGENASE | | Authors: | Dhindwal, S, Patil, D.N, Kumar, P. | | Deposit date: | 2011-07-23 | | Release date: | 2011-08-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Biochemical Studies and Ligand-Bound Structures of Biphenyl Dehydrogenase from Pandoraea Pnomenusa Strain B-356 Reveal a Basis for Broad Specificity of the Enzyme.
J.Biol.Chem., 286, 2011
|
|
7ZC9
 
 | | Human Pikachurin/EGFLAM C-terminal Laminin-G domain (LG3) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Pikachurin, SULFATE ION | | Authors: | Pantalone, S, Forneris, F. | | Deposit date: | 2022-03-26 | | Release date: | 2023-07-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the photoreceptor synaptic assembly of the extracellular matrix protein pikachurin with the orphan receptor GPR179.
Sci.Signal., 16, 2023
|
|
7ZCB
 
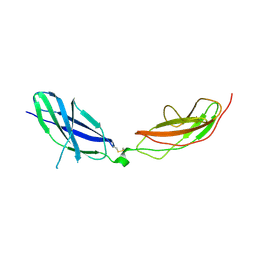 | | Human Pikachurin/EGFLAM N-terminal Fibronectin-III (1-2) domains | | Descriptor: | CHLORIDE ION, Pikachurin, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Pantalone, S, Savino, S, Viti, L.V, Forneris, F. | | Deposit date: | 2022-03-26 | | Release date: | 2023-07-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the photoreceptor synaptic assembly of the extracellular matrix protein pikachurin with the orphan receptor GPR179.
Sci.Signal., 16, 2023
|
|
4UMD
 
 | |
4UM7
 
 | | Crystal structure of 3-deoxy-D-manno-octulosonate 8-phosphate phosphatase (kdsC) from Moraxella catarrhalis in complex with Magnesium ion | | Descriptor: | 1,2-ETHANEDIOL, 3-DEOXY-D-MANNO-OCTULOSONATE 8-PHOSPHATE PHOSPHATASE KDSC, MAGNESIUM ION | | Authors: | Dhindwal, S, Tomar, S, Kumar, P. | | Deposit date: | 2014-05-15 | | Release date: | 2015-02-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Ligand-Bound Structures of 3-Deoxy-D-Manno-Octulosonate 8-Phosphate Phosphatase from Moraxella Catarrhalis Reveal a Water Channel Connecting to the Active Site for the Second Step of Catalysis
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4UM5
 
 | | Crystal structure of 3-deoxy-D-manno-octulosonate 8-phosphate phosphatase from Moraxella catarrhalis in complex with Magnesium ion and Phosphate ion | | Descriptor: | 1,2-ETHANEDIOL, 3-DEOXY-D-MANNO-OCTULOSONATE 8-PHOSPHATE PHOSPHATASE KDSC, MAGNESIUM ION, ... | | Authors: | Dhindwal, S, Tomar, S, Kumar, P. | | Deposit date: | 2014-05-15 | | Release date: | 2015-02-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Ligand-Bound Structures of 3-Deoxy-D-Manno-Octulosonate 8-Phosphate Phosphatase from Moraxella Catarrhalis Reveal a Water Channel Connecting to the Active Site for the Second Step of Catalysis
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4UMF
 
 | | Crystal structure of 3-deoxy-D-manno-octulosonate 8-phosphate phosphatase from Moraxella catarrhalis in complex with Magnesium ion, Phosphate ion and KDO molecule | | Descriptor: | 3-DEOXY-D-MANNO-OCTULOSONATE 8-PHOSPHATE PHOSPHATASE KDSC, 3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid, MAGNESIUM ION, ... | | Authors: | Dhindwal, S, Tomar, S, Kumar, P. | | Deposit date: | 2014-05-16 | | Release date: | 2015-02-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Ligand-Bound Structures of 3-Deoxy-D-Manno-Octulosonate 8-Phosphate Phosphatase from Moraxella Catarrhalis Reveal a Water Channel Connecting to the Active Site for the Second Step of Catalysis
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4UME
 
 | | Crystal structure of 3-deoxy-D-manno-octulosonate 8-phosphate phosphatase from Moraxella catarrhalis in complex with Magnesium ion and KDO molecule | | Descriptor: | 3-DEOXY-D-MANNO-OCTULOSONATE 8-PHOSPHATE PHOSPHATASE KDSC, 3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid, MAGNESIUM ION | | Authors: | Dhindwal, S, Tomar, S, Kumar, P. | | Deposit date: | 2014-05-16 | | Release date: | 2015-02-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Ligand-Bound Structures of 3-Deoxy-D-Manno-Octulosonate 8-Phosphate Phosphatase from Moraxella Catarrhalis Reveal a Water Channel Connecting to the Active Site for the Second Step of Catalysis
Acta Crystallogr.,Sect.D, 71, 2015
|
|
7LWH
 
 | | Human neurofibromin 2/merlin residues 1-339 in complex with LATS1 | | Descriptor: | GLYCEROL, IMIDAZOLE, Merlin, ... | | Authors: | Primi, M.C, Rangarajan, E.S, Izard, T. | | Deposit date: | 2021-03-01 | | Release date: | 2021-08-11 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.606 Å) | | Cite: | Conformational flexibility determines the Nf2/merlin tumor suppressor functions.
Matrix Biol Plus, 12, 2021
|
|
