1COL
 
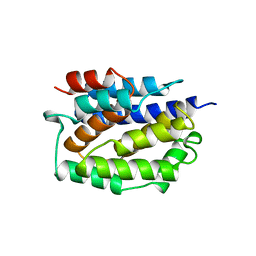 | | REFINED STRUCTURE OF THE PORE-FORMING DOMAIN OF COLICIN A AT 2.4 ANGSTROMS RESOLUTION | | Descriptor: | COLICIN A | | Authors: | Parker, M.W, Postma, J.P.M, Pattus, F, Tucker, A.D, Tsernoglou, D. | | Deposit date: | 1991-07-06 | | Release date: | 1992-07-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Refined structure of the pore-forming domain of colicin A at 2.4 A resolution.
J.Mol.Biol., 224, 1992
|
|
1PRE
 
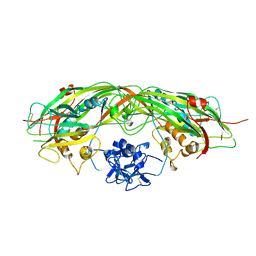 | | PROAEROLYSIN | | Descriptor: | PROAEROLYSIN | | Authors: | Parker, M.W, Buckley, J.T, Postma, J.P.M, Tucker, A.D, Tsernoglou, D. | | Deposit date: | 1995-09-15 | | Release date: | 1996-10-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the Aeromonas toxin proaerolysin in its water-soluble and membrane-channel states.
Nature, 367, 1994
|
|
4QDQ
 
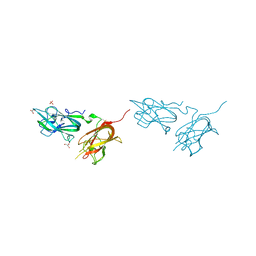 | | Physical basis for Nrp2 ligand binding | | Descriptor: | GLYCEROL, Neuropilin-2, SULFATE ION | | Authors: | Parker, M.W, Vander Kooi, C.W. | | Deposit date: | 2014-05-14 | | Release date: | 2015-04-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Basis for VEGF-C Binding to Neuropilin-2 and Sequestration by a Soluble Splice Form.
Structure, 23, 2015
|
|
4QDR
 
 | |
4QDS
 
 | | Physical basis for Nrp2 ligand binding | | Descriptor: | ACETATE ION, GLYCEROL, Neuropilin-2 | | Authors: | Parker, M.W, Vander Kooi, C.W. | | Deposit date: | 2014-05-14 | | Release date: | 2015-04-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis for VEGF-C Binding to Neuropilin-2 and Sequestration by a Soluble Splice Form.
Structure, 23, 2015
|
|
1Z52
 
 | | Proaerolysin Mutant W373L | | Descriptor: | Aerolysin | | Authors: | Parker, M.W, Feil, S.C, Tang, J.W. | | Deposit date: | 2005-03-16 | | Release date: | 2006-03-07 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Crystal Structure of Proaerolysin at 2.3 A Resolution and Structural Analyses of Single-site Mutants as a Basis for Understanding Membrane Insertion of the Toxin
To be Published
|
|
5DHL
 
 | |
5DIM
 
 | |
4ZGH
 
 | | Structure of Sugar Binding Protein Pneumolysin | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GOLD (I) CYANIDE ION, ... | | Authors: | Parker, M.W, Feil, S.C, Morton, C. | | Deposit date: | 2015-04-23 | | Release date: | 2016-03-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of Streptococcus pneumoniae pneumolysin provides key insights into early steps of pore formation.
Sci Rep, 5, 2015
|
|
4NKQ
 
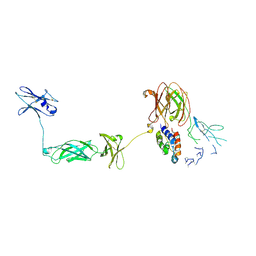 | | Structure of a Cytokine Receptor Complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cytokine receptor common subunit beta, Granulocyte-macrophage colony-stimulating factor, ... | | Authors: | Parker, M.W, Broughton, S.E. | | Deposit date: | 2013-11-13 | | Release date: | 2015-09-23 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.301 Å) | | Cite: | Conformational Changes in the GM-CSF Receptor Suggest a Molecular Mechanism for Affinity Conversion and Receptor Signaling.
Structure, 24, 2016
|
|
1GSS
 
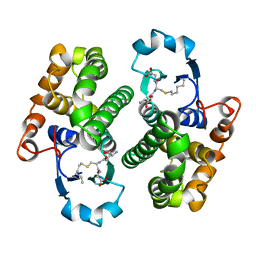 | | THREE-DIMENSIONAL STRUCTURE OF CLASS PI GLUTATHIONE S-TRANSFERASE FROM HUMAN PLACENTA IN COMPLEX WITH S-HEXYLGLUTATHIONE AT 2.8 ANGSTROMS RESOLUTION | | Descriptor: | GLUTATHIONE S-TRANSFERASE, L-gamma-glutamyl-S-hexyl-L-cysteinylglycine | | Authors: | Reinemer, P, Dirr, H.W, Ladenstein, R, Lobello, M, Federici, G, Huber, R, Parker, M.W. | | Deposit date: | 1992-05-28 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Three-dimensional structure of class pi glutathione S-transferase from human placenta in complex with S-hexylglutathione at 2.8 A resolution.
J.Mol.Biol., 227, 1992
|
|
6PRO
 
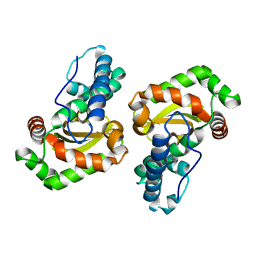 | | MnSOD from Geobacillus stearothermophilus | | Descriptor: | MANGANESE (II) ION, Superoxide dismutase | | Authors: | Adams, J.J, Morton, C.J, Parker, M.W. | | Deposit date: | 2019-07-10 | | Release date: | 2020-05-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.263 Å) | | Cite: | The Crystal Structure of the Manganese Superoxide Dismutase from Geobacillus stearothermophilus: Parker and Blake (1988) Revisited
Aust.J.Chem., 73, 2020
|
|
2LJR
 
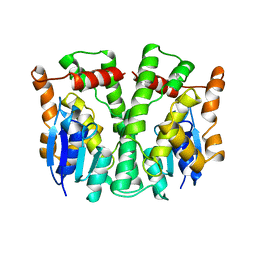 | | GLUTATHIONE TRANSFERASE APO-FORM FROM HUMAN | | Descriptor: | GLUTATHIONE S-TRANSFERASE | | Authors: | Rossjohn, J, Mckinstry, W.J, Oakley, A.J, Verger, D, Flanagan, J, Chelvanayagam, G, Tan, K.L, Board, P.G, Parker, M.W. | | Deposit date: | 1998-03-08 | | Release date: | 1999-03-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Human theta class glutathione transferase: the crystal structure reveals a sulfate-binding pocket within a buried active site.
Structure, 6, 1998
|
|
2A2R
 
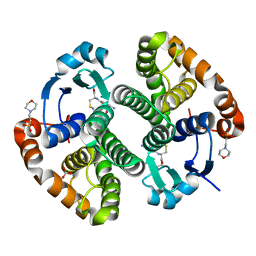 | | Crystal Structure of Glutathione Transferase Pi in complex with S-nitrosoglutathione | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-AMINO-5-[1-(CARBOXYLATOMETHYLCARBAMOYL)-2-NITROSOSULFANYL-ETHYL]AMINO-5-OXO-PENTANOATE, CALCIUM ION, ... | | Authors: | Parker, L.J, Morton, C.J, Adams, J.J, Parker, M.W. | | Deposit date: | 2005-06-23 | | Release date: | 2006-06-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Calorimetric and structural studies of the nitric oxide carrier S-nitrosoglutathione bound to human glutathione transferase P1-1
Protein Sci., 15, 2006
|
|
2A2S
 
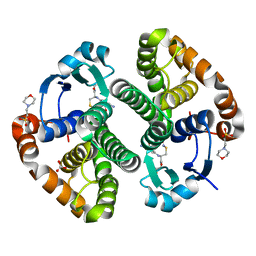 | | Crystal Structure of Human Glutathione Transferase in complex with S-nitrosoglutathione in the absence of reducing agent | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-AMINO-5-[1-(CARBOXYLATOMETHYLCARBAMOYL)-2-NITROSOSULFANYL-ETHYL]AMINO-5-OXO-PENTANOATE, CALCIUM ION, ... | | Authors: | Parker, L.J, Morton, C.J, Adams, J.J, Parker, M.W. | | Deposit date: | 2005-06-23 | | Release date: | 2006-06-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Calorimetric and structural studies of the nitric oxide carrier S-nitrosoglutathione bound to human glutathione transferase P1-1
Protein Sci., 15, 2006
|
|
2AEW
 
 | |
6PDG
 
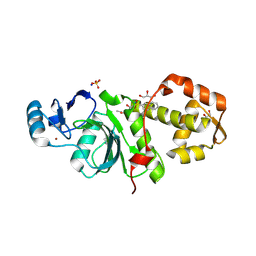 | | Crystal structure of MYST acetyltransferase domain in complex with inhibitor 83 | | Descriptor: | 5-ethoxy-2-fluoro-3-methyl-N'-[(naphthalen-2-yl)sulfonyl]benzohydrazide, GLYCEROL, Histone acetyltransferase KAT8, ... | | Authors: | Hermans, S.J, Parker, M.W, Thomas, T, Baell, J.B. | | Deposit date: | 2019-06-18 | | Release date: | 2020-04-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.919 Å) | | Cite: | Discovery of Acylsulfonohydrazide-Derived Inhibitors of the Lysine Acetyltransferase, KAT6A, as Potent Senescence-Inducing Anti-Cancer Agents.
J.Med.Chem., 63, 2020
|
|
6PDA
 
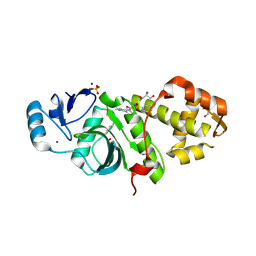 | | Crystal structure of MYST acetyltransferase domain in complex with inhibitor 74 | | Descriptor: | 2-fluoro-3-methyl-N'-(phenylsulfonyl)-5-(pyridin-2-yl)benzohydrazide, GLYCEROL, Histone acetyltransferase KAT8, ... | | Authors: | Hermans, S.J, Parker, M.W, Thomas, T, Baell, J.B. | | Deposit date: | 2019-06-18 | | Release date: | 2020-04-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.448 Å) | | Cite: | Discovery of Acylsulfonohydrazide-Derived Inhibitors of the Lysine Acetyltransferase, KAT6A, as Potent Senescence-Inducing Anti-Cancer Agents.
J.Med.Chem., 63, 2020
|
|
6PDE
 
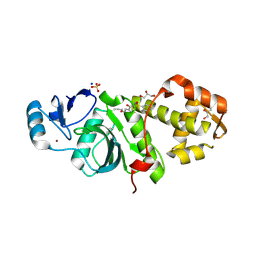 | | Crystal structure of MYST acetyltransferase domain in complex with inhibitor 40 | | Descriptor: | 2-fluoro-3-methyl-N'-(phenylsulfonyl)-5-propoxybenzohydrazide, GLYCEROL, Histone acetyltransferase KAT8, ... | | Authors: | Hermans, S.J, Parker, M.W, Thomas, T, Baell, J.B. | | Deposit date: | 2019-06-18 | | Release date: | 2020-04-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Discovery of Acylsulfonohydrazide-Derived Inhibitors of the Lysine Acetyltransferase, KAT6A, as Potent Senescence-Inducing Anti-Cancer Agents.
J.Med.Chem., 63, 2020
|
|
8G32
 
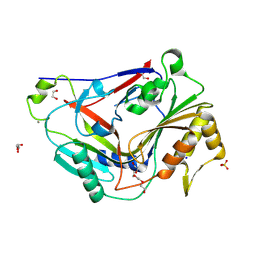 | | Pro-form of a CDCL short from E. anophelis | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, GLYCEROL, ... | | Authors: | Johnstone, B.A, Christie, M.P, Morton, C.J, Parker, M.W. | | Deposit date: | 2023-02-06 | | Release date: | 2024-02-07 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Distant relatives of a eukaryotic cell-specific toxin family evolved a complement-like mechanism to kill bacteria.
Nat Commun, 15, 2024
|
|
8G33
 
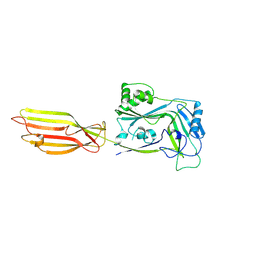 | |
4Y0G
 
 | | beta2 carbohydrate binding module (CBM) of AMP-activated protein kinase (AMPK) | | Descriptor: | 5'-AMP-activated protein kinase subunit beta-2, GLYCEROL | | Authors: | Mobbs, J, Gorman, M.A, Parker, M.W, Gooley, P.R, Griffin, M. | | Deposit date: | 2015-02-06 | | Release date: | 2015-04-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Determinants of oligosaccharide specificity of the carbohydrate-binding modules of AMP-activated protein kinase.
Biochem.J., 468, 2015
|
|
4XXD
 
 | | Crystal Structure of mid-region amyloid beta capture by solanezumab | | Descriptor: | Amyloid-beta fragment, Fab Heavy Chain, Fab Light Chain | | Authors: | Hermans, S.J, Crespi, G.A.N, Parker, M.W, Miles, L.A. | | Deposit date: | 2015-01-30 | | Release date: | 2015-04-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Molecular basis for mid-region amyloid-beta capture by leading Alzheimer's disease immunotherapies.
Sci Rep, 5, 2015
|
|
6BFQ
 
 | | The mechanism of GM-CSF inhibition by human GM-CSF auto-antibodies | | Descriptor: | Fab Heavy chain, Fab Light Chain, Granulocyte-macrophage colony-stimulating factor | | Authors: | Dhagat, U, Hercus, T.R, Broughton, S.E, Nero, T.L, Lopez, A.F, Parker, M.W. | | Deposit date: | 2017-10-26 | | Release date: | 2018-09-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The mechanism of GM-CSF inhibition by human GM-CSF auto-antibodies suggests novel therapeutic opportunities.
MAbs, 10, 2018
|
|
1S3R
 
 | | Crystal structure of the human-specific toxin intermedilysin | | Descriptor: | SULFATE ION, intermedilysin | | Authors: | Polekhina, G, Giddings, K.S, Tweten, R.K, Parker, M.W. | | Deposit date: | 2004-01-14 | | Release date: | 2005-01-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Insights into the action of the superfamily of cholesterol-dependent cytolysins from studies of intermedilysin
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
