1L1G
 
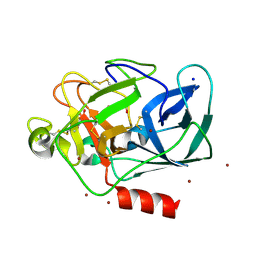 | | The Structure of Porcine Pancreatic Elastase Complexed with Xenon and Bromide, Cryoprotected with Glycerol | | Descriptor: | BROMIDE ION, ELASTASE 1, GLYCEROL, ... | | Authors: | Panjikar, S, Tucker, P.A. | | Deposit date: | 2002-02-16 | | Release date: | 2002-08-28 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Xenon derivatization of halide-soaked protein crystals.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1L0Z
 
 | | THE STRUCTURE OF PORCINE PANCREATIC ELASTASE COMPLEXED WITH XENON AND BROMIDE, CRYOPROTECTED WITH DRY PARAFFIN OIL | | Descriptor: | BROMIDE ION, ELASTASE 1, SODIUM ION, ... | | Authors: | Tucker, P.A, Panjikar, S. | | Deposit date: | 2002-02-14 | | Release date: | 2002-08-28 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Xenon derivatization of halide-soaked protein crystals.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1URJ
 
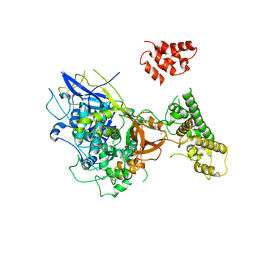 | | Single stranded DNA-binding protein(ICP8) from Herpes simplex virus-1 | | Descriptor: | MAJOR DNA-BINDING PROTEIN, MERCURY (II) ION, ZINC ION | | Authors: | Panjikar, S, Mapelli, M, Tucker, P.A. | | Deposit date: | 2003-10-30 | | Release date: | 2004-11-11 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The crystal structure of the herpes simplex virus 1 ssDNA-binding protein suggests the structural basis for flexible, cooperative single-stranded DNA binding.
J. Biol. Chem., 280, 2005
|
|
2FPB
 
 | |
2FP8
 
 | |
2FPC
 
 | |
2FP9
 
 | | Crystal structure of Native Strictosidine Synthase | | Descriptor: | L(+)-TARTARIC ACID, Strictosidine synthase | | Authors: | Panjikar, S. | | Deposit date: | 2006-01-16 | | Release date: | 2006-05-23 | | Last modified: | 2019-07-10 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | The structure of Rauvolfia serpentina strictosidine synthase is a novel six-bladed beta-propeller fold in plant proteins
Plant Cell, 18, 2006
|
|
7MDS
 
 | | Crystal structure of AtDHDPS1 in complex with MBDTA-2 | | Descriptor: | 4-hydroxy-tetrahydrodipicolinate synthase 1, chloroplastic, CHLORIDE ION, ... | | Authors: | Hall, C.J, Soares da Costa, T.P, Panjikar, S. | | Deposit date: | 2021-04-06 | | Release date: | 2021-08-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.295 Å) | | Cite: | Towards novel herbicide modes of action by inhibiting lysine biosynthesis in plants.
Elife, 10, 2021
|
|
1O1H
 
 | | STRUCTURE OF GLUCOSE ISOMERASE DERIVATIZED WITH KR. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, ... | | Authors: | Nowak, E, Panjikar, S, Tucker, P.A. | | Deposit date: | 2002-11-07 | | Release date: | 2002-11-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of Glucose Isomerase Derivatized with Kr.
To be Published
|
|
8D4Y
 
 | | C-terminal SANT-SLIDE domain of human Chromodomain-helicase-DNA-binding protein 4 (CHD4) | | Descriptor: | Chromodomain-helicase-DNA-binding protein 4 | | Authors: | Moghaddas Sani, H, Deshpande, C.N, Panjikar, S, Patel, K, Mackay, J.P. | | Deposit date: | 2022-06-03 | | Release date: | 2022-12-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The role of auxiliary domains in modulating CHD4 activity suggests mechanistic commonality between enzyme families.
Nat Commun, 13, 2022
|
|
5KBI
 
 | | CRYSTAL STRUCTURE OF THE AROMATIC SENSOR DOMAIN OF MOPR IN COMPLEX WITH CATACHOL | | Descriptor: | CATECHOL, MopR, ZINC ION | | Authors: | Ray, S, Anand, R, Panjikar, S. | | Deposit date: | 2016-06-03 | | Release date: | 2016-07-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural Basis of Selective Aromatic Pollutant Sensing by the Effector Binding Domain of MopR, an NtrC Family Transcriptional Regulator.
Acs Chem.Biol., 11, 2016
|
|
7N3V
 
 | |
8E7F
 
 | | Crystal structure of the autotransporter Ssp from Serratia marcescens. | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, Extracellular serine protease, ... | | Authors: | Hor, L, Pilapitiya, A, Panjikar, S, Paxman, J.J, Heras, B. | | Deposit date: | 2022-08-23 | | Release date: | 2023-03-08 | | Last modified: | 2023-03-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a subtilisin-like autotransporter passenger domain reveals insights into its cytotoxic function.
Nat Commun, 14, 2023
|
|
6EBB
 
 | |
5KBG
 
 | | CRYSTAL STRUCTURE OF THE AROMATIC SENSOR DOMAIN OF MOPR IN COMPLEX WITH OCRESOL | | Descriptor: | MopR, ZINC ION, o-cresol | | Authors: | Ray, S, Anand, R, Panjikar, S. | | Deposit date: | 2016-06-03 | | Release date: | 2016-07-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis of Selective Aromatic Pollutant Sensing by the Effector Binding Domain of MopR, an NtrC Family Transcriptional Regulator.
Acs Chem.Biol., 11, 2016
|
|
5KBE
 
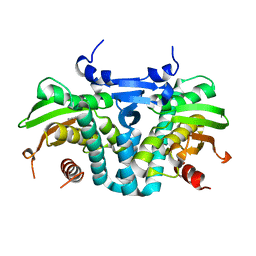 | | CRYSTAL STRUCTURE OF THE AROMATIC SENSOR DOMAIN OF MOPR IN COMPLEX WITH PHENOL | | Descriptor: | MopR, PHENOL, ZINC ION | | Authors: | Ray, S, Anand, R, Panjikar, S. | | Deposit date: | 2016-06-03 | | Release date: | 2016-07-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis of Selective Aromatic Pollutant Sensing by the Effector Binding Domain of MopR, an NtrC Family Transcriptional Regulator.
Acs Chem.Biol., 11, 2016
|
|
5KBH
 
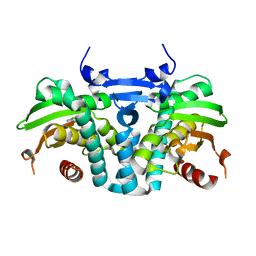 | | CRYSTAL STRUCTURE OF THE AROMATIC SENSOR DOMAIN OF MOPR IN COMPLEX WITH 3-CHLORO-PHENOL | | Descriptor: | 3-CHLOROPHENOL, MopR, ZINC ION | | Authors: | Ray, S, Anand, R, Panjikar, S. | | Deposit date: | 2016-06-03 | | Release date: | 2016-07-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural Basis of Selective Aromatic Pollutant Sensing by the Effector Binding Domain of MopR, an NtrC Family Transcriptional Regulator.
Acs Chem.Biol., 11, 2016
|
|
4WVM
 
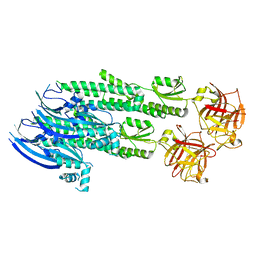 | | Stonustoxin structure | | Descriptor: | Stonustoxin subunit alpha, Stonustoxin subunit beta | | Authors: | Ellisdon, A.M, Panjikar, S, Whisstock, J.C, McGowan, S. | | Deposit date: | 2014-11-06 | | Release date: | 2015-12-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Stonefish toxin defines an ancient branch of the perforin-like superfamily.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5E5T
 
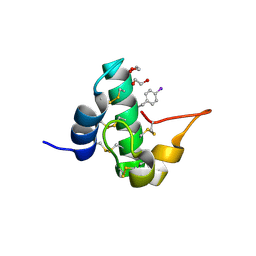 | | Quasi-racemic snakin-1 in P1 after radiation damage | | Descriptor: | 1,2-ETHANEDIOL, D- snakin-1, FORMIC ACID, ... | | Authors: | Yeung, H, Squire, C.J, Yosaatmadja, Y, Panjikar, S, Baker, E.N, Harris, P.W.R, Brimble, M.A. | | Deposit date: | 2015-10-09 | | Release date: | 2016-05-18 | | Last modified: | 2016-07-20 | | Method: | X-RAY DIFFRACTION (1.572 Å) | | Cite: | Radiation Damage and Racemic Protein Crystallography Reveal the Unique Structure of the GASA/Snakin Protein Superfamily.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
5E5Q
 
 | | Racemic snakin-1 in P21/c | | Descriptor: | Snakin-1 | | Authors: | Yeung, H, Squire, C.J, Yosaatmadja, Y, Panjikar, S, Baker, E.N, Harris, P.W.R, Brimble, M.A. | | Deposit date: | 2015-10-09 | | Release date: | 2016-05-18 | | Last modified: | 2016-07-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Radiation Damage and Racemic Protein Crystallography Reveal the Unique Structure of the GASA/Snakin Protein Superfamily.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
5E5Y
 
 | | Quasi-racemic snakin-1 in P1 before radiation damage | | Descriptor: | 1,2-ETHANEDIOL, D- snakin-1, FORMIC ACID, ... | | Authors: | Yeung, H, Squire, C.J, Yosaatmadja, Y, Panjikar, S, Baker, E.N, Harris, P.W.R, Brimble, M.A. | | Deposit date: | 2015-10-09 | | Release date: | 2016-05-18 | | Last modified: | 2016-07-20 | | Method: | X-RAY DIFFRACTION (1.506 Å) | | Cite: | Radiation Damage and Racemic Protein Crystallography Reveal the Unique Structure of the GASA/Snakin Protein Superfamily.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
4GUV
 
 | | TetX derivatized with Xenon | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION, TetX2 protein, ... | | Authors: | Volkers, G, Palm, G.J, Panjikar, S, Hinrichs, W. | | Deposit date: | 2012-08-29 | | Release date: | 2013-04-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Putative dioxygen-binding sites and recognition of tigecycline and minocycline in the tetracycline-degrading monooxygenase TetX.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4IGL
 
 | | Structure of the RHS-repeat containing BC component of the secreted ABC toxin complex from Yersinia entomophaga | | Descriptor: | GLYCEROL, POTASSIUM ION, YenB, ... | | Authors: | Busby, J.N, Panjikar, S, Landsberg, M.J, Hurst, M.R.H, Lott, J.S. | | Deposit date: | 2012-12-17 | | Release date: | 2013-08-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | The BC component of ABC toxins is an RHS-repeat-containing protein encapsulation device
Nature, 501, 2013
|
|
5KT0
 
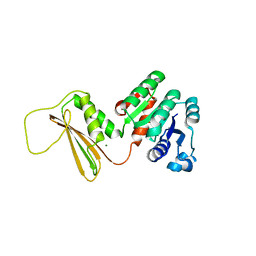 | | Dihydrodipicolinate reductase from the industrial and evolutionarily important cyanobacteria Anabaena variabilis. | | Descriptor: | 4-hydroxy-tetrahydrodipicolinate reductase, MAGNESIUM ION | | Authors: | Christensen, J.B, Soares da Costa, T.P, Faou, P, Pearce, F.G, Panjikar, S, Perugini, M.A. | | Deposit date: | 2016-07-10 | | Release date: | 2016-09-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Dihydrodipicolinate Reductase from the industrial and evolutionarily important cyanobacteria Anabaena variabilis.
To Be Published
|
|
5KTL
 
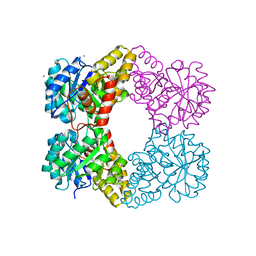 | |
