5FOJ
 
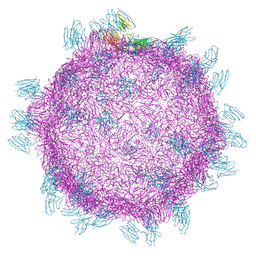 | | Cryo electron microscopy structure of Grapevine Fanleaf Virus complex with Nanobody | | Descriptor: | Nanobody, RNA2 polyprotein | | Authors: | Orlov, I, Hemmer, C, Ackerer, L, Lorber, B, Ghannam, A, Poignavent, V, Hleibieh, K, Sauter, C, Schmitt-Keichinger, C, Belval, L, Hily, J.M, Marmonier, A, Komar, V, Gersch, S, Schellenberger, P, Bron, P, Vigne, E, Muyldermans, S, Lemaire, O, Demangeat, G, Ritzenthaler, C, Klaholz, B.P. | | Deposit date: | 2015-11-22 | | Release date: | 2016-01-20 | | Last modified: | 2021-08-11 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis of nanobody recognition of grapevine fanleaf virus and of virus resistance loss.
Proc.Natl.Acad.Sci.USA, 2020
|
|
7Z4W
 
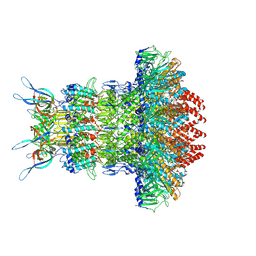 | | gp6/gp15/gp16 connector complex of bacteriophage SPP1 | | Descriptor: | Head completion protein gp15, Head completion protein gp16, Portal protein | | Authors: | Orlov, I, Roche, S, Tavares, P, Orlova, E.V. | | Deposit date: | 2022-03-05 | | Release date: | 2022-11-30 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | CryoEM structure and assembly mechanism of a bacterial virus genome gatekeeper.
Nat Commun, 13, 2022
|
|
4L99
 
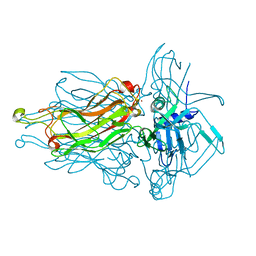 | | Structure of the RBP from lactococcal phage 1358 in complex with glycerol | | Descriptor: | GLYCEROL, Receptor Binding Protein, ZINC ION | | Authors: | Farenc, C, Spinelli, S, Bebeacua, C, Tremblay, D, Orlov, I, Blangy, S, Klaholz, B.P, Moineau, S, Cambillau, C. | | Deposit date: | 2013-06-18 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A Virulent Siphophage CyoEM Structure and Host Recognition and Infection Mechanism
To be Published
|
|
4UMM
 
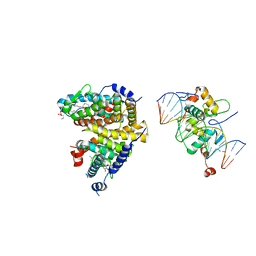 | | The Cryo-EM structure of the palindromic DNA-bound USP-EcR nuclear receptor reveals an asymmetric organization with allosteric domain positioning | | Descriptor: | 2,3,14,20,22-PENTAHYDROXYCHOLEST-7-EN-6-ONE, 5'-D(*CP*AP*AP*GP*GP*GP*TP*TP*CP*AP*AP*TP*GP*CP *AP*CP*TP*TP*GP*TP)-3', 5'-D(*DGP*AP*CP*AP*AP*GP*TP*GP*CP*AP*TP*TP*GP*DAP *AP*CP*CP*CP*TP*T)-3', ... | | Authors: | Maletta, M, Orlov, I, Moras, D, Billas, I.M.L, Klaholz, B.P. | | Deposit date: | 2014-05-19 | | Release date: | 2014-06-25 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (11.6 Å) | | Cite: | The Palindromic DNA-Bound Usp-Ecr Nuclear Receptor Adopts an Asymmetric Organization with Allosteric Domain Positioning.
Nat.Commun., 5, 2014
|
|
4L97
 
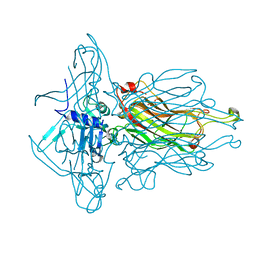 | | Structure of the RBP of lactococcal phage 1358 in complex with glucose-1-phosphate | | Descriptor: | 1-O-phosphono-alpha-D-glucopyranose, Receptor Binding Protein | | Authors: | Farenc, C, Spinelli, S, Bebeacua, C, Tremblay, D, Orlov, I, Blangy, S, Klaholz, B.P, Moineau, S, Cambillau, C. | | Deposit date: | 2013-06-18 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | A Virulent Siphophage CyoEM Structure and Host Recognition and Infection Mechanism
To be Published
|
|
4L92
 
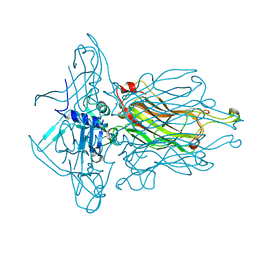 | | Structure of the RBP from lactococcal phage 1358 in complex with 2 GlcNAc molecules | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, Receptor Binding Protein, ... | | Authors: | Farenc, C, Spinelli, S, Bebeacua, C, Tremblay, D, Orlov, I, Blangy, S, Klaholz, B.P, Moineau, S, Cambillau, C. | | Deposit date: | 2013-06-18 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Virulent Siphophage CyoEM Structure and Host Recognition and Infection Mechanism
To be Published
|
|
4L9B
 
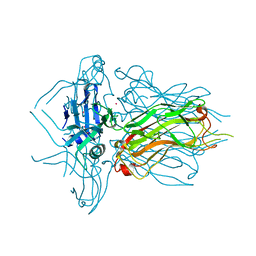 | | Structure of native RBP from lactococcal phage 1358 (CsI derivative) | | Descriptor: | CESIUM ION, Receptor Binding Protein | | Authors: | Farenc, C, Spinelli, S, Bebeacua, C, Tremblay, D, Orlov, I, Blangy, S, Klaholz, B.P, Moineau, S, Cambillau, C. | | Deposit date: | 2013-06-18 | | Release date: | 2014-04-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A Virulent Siphophage CyoEM Structure and Host Recognition and Infection Mechanism
To be Published
|
|
4MKQ
 
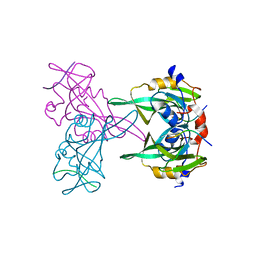 | |
4MJT
 
 | |
4MKO
 
 | |
5OI8
 
 | |
5OIA
 
 | |
5OI2
 
 | |
5OI5
 
 | |
5OI3
 
 | |
3C1O
 
 | |
3C3X
 
 | |
