2ZUK
 
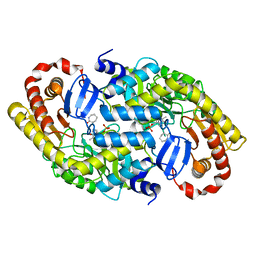 | | The crystal structure of alpha-amino-epsilon-caprolactam racemase from Achromobacter obae complexed with epsilon caprolactam (different binding mode) | | Descriptor: | Alpha-amino-epsilon-caprolactam racemase, PYRIDOXAL-5'-PHOSPHATE, azepan-2-one | | Authors: | Okazaki, S, Suzuki, A, Komeda, H, Asano, Y, Yamane, T. | | Deposit date: | 2008-10-18 | | Release date: | 2009-02-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | The novel structure of a pyridoxal 5'-phosphate-dependent fold-type I racemase, alpha-amino-epsilon-caprolactam racemase from Achromobacter obae
Biochemistry, 48, 2009
|
|
2DCN
 
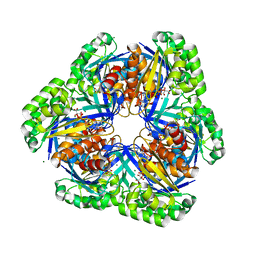 | | Crystal structure of 2-keto-3-deoxygluconate kinase from Sulfolobus tokodaii complexed with 2-keto-6-phosphogluconate (alpha-furanose form) | | Descriptor: | 6-O-phosphono-beta-D-psicofuranosonic acid, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Okazaki, S, Onda, H, Suzuki, A, Kuramitsu, S, Masui, R, Yamane, T. | | Deposit date: | 2006-01-10 | | Release date: | 2006-01-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of 2-keto-3-deoxygluconate kinase from Sulfolobus tokodaii complexed with 2-keto-6-phosphogluconate
To be Published
|
|
2DRW
 
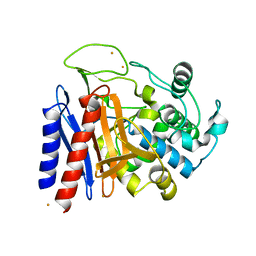 | | The crystal structutre of D-amino acid amidase from Ochrobactrum anthropi SV3 | | Descriptor: | BARIUM ION, D-Amino acid amidase | | Authors: | Okazaki, S, Suzuki, A, Komeda, H, Asano, Y, Yamane, T. | | Deposit date: | 2006-06-15 | | Release date: | 2006-07-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure and Functional Characterization of a D-Stereospecific Amino Acid Amidase from Ochrobactrum anthropi SV3, a New Member of the Penicillin-recognizing Proteins
J.Mol.Biol., 368, 2007
|
|
2DNS
 
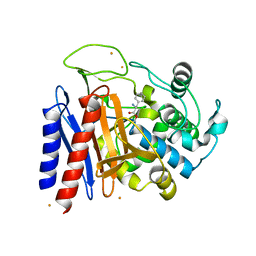 | | The crystal structure of D-amino acid amidase from Ochrobactrum anthropi SV3 complexed with D-Phenylalanine | | Descriptor: | BARIUM ION, D-PHENYLALANINE, D-amino acid amidase | | Authors: | Okazaki, S, Suzuki, A, Komeda, H, Asano, Y, Yamane, T. | | Deposit date: | 2006-04-26 | | Release date: | 2006-05-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure and Functional Characterization of a D-Stereospecific Amino Acid Amidase from Ochrobactrum anthropi SV3, a New Member of the Penicillin-recognizing Proteins
J.Mol.Biol., 368, 2007
|
|
3DXW
 
 | | The crystal structure of alpha-amino-epsilon-caprolactam racemase from Achromobacter obae complexed with epsilon caprolactam | | Descriptor: | Alpha-amino-epsilon-caprolactam racemase, PYRIDOXAL-5'-PHOSPHATE, azepan-2-one | | Authors: | Okazaki, S, Suzuki, A, Komeda, H, Asano, Y, Yamane, T. | | Deposit date: | 2008-07-25 | | Release date: | 2009-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | The novel structure of a pyridoxal 5'-phosphate-dependent fold-type I racemase, alpha-amino-epsilon-caprolactam racemase from Achromobacter obae
Biochemistry, 48, 2009
|
|
3DXV
 
 | | The crystal structure of alpha-amino-epsilon-caprolactam racemase from Achromobacter obae | | Descriptor: | Alpha-amino-epsilon-caprolactam racemase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Okazaki, S, Suzuki, A, Komeda, H, Asano, Y, Yamane, T. | | Deposit date: | 2008-07-25 | | Release date: | 2009-02-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | The novel structure of a pyridoxal 5'-phosphate-dependent fold-type I racemase, alpha-amino-epsilon-caprolactam racemase from Achromobacter obae
Biochemistry, 48, 2009
|
|
3WEV
 
 | | Crystal structure of the Schiff base intermediate of L-Lys epsilon-oxidase from Marinomonas mediterranea with L-Lys | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIETHYLENE DIOXIDE, L-lysine 6-oxidase, ... | | Authors: | Okazaki, S, Nakano, S, Matsui, D, Akaji, S, Inagaki, K, Asano, Y. | | Deposit date: | 2013-07-12 | | Release date: | 2013-09-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | X-Ray crystallographic evidence for the presence of the cysteine tryptophylquinone cofactor in L-lysine {varepsilon}-oxidase from Marinomonas mediterranea
J.Biochem., 154, 2013
|
|
3WEU
 
 | | Crystal structure of the L-Lys epsilon-oxidase from Marinomonas mediterranea | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIETHYLENE DIOXIDE, L-lysine 6-oxidase, ... | | Authors: | Okazaki, S, Nakano, S, Matsui, D, Akaji, S, Inagaki, K, Asano, Y. | | Deposit date: | 2013-07-12 | | Release date: | 2013-09-04 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | X-Ray crystallographic evidence for the presence of the cysteine tryptophylquinone cofactor in L-lysine {varepsilon}-oxidase from Marinomonas mediterranea
J.Biochem., 154, 2013
|
|
2EFX
 
 | | The crystal structure of D-amino acid amidase from Ochrobactrum anthropi SV3 complexed with L-phenylalanine amide | | Descriptor: | BARIUM ION, D-amino acid amidase, PHENYLALANINE AMIDE | | Authors: | Okazaki, S, Suzuki, A, Mizushima, T, Komeda, H, Asano, Y, Yamane, T. | | Deposit date: | 2007-02-26 | | Release date: | 2007-03-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of D-amino-acid amidase complexed with L-phenylalanine and with L-phenylalanine amide: insight into the D-stereospecificity of D-amino-acid amidase from Ochrobactrum anthropi SV3.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
2EFU
 
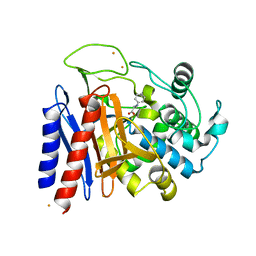 | | The crystal structure of D-amino acid amidase from Ochrobactrum anthropi SV3 complexed with L-phenylalanine | | Descriptor: | BARIUM ION, D-Amino acid amidase, PHENYLALANINE | | Authors: | Okazaki, S, Suzuki, A, Mizushima, T, Komeda, H, Asano, Y, Yamane, T. | | Deposit date: | 2007-02-26 | | Release date: | 2007-03-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of D-amino-acid amidase complexed with L-phenylalanine and with L-phenylalanine amide: insight into the D-stereospecificity of D-amino-acid amidase from Ochrobactrum anthropi SV3.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
3VIA
 
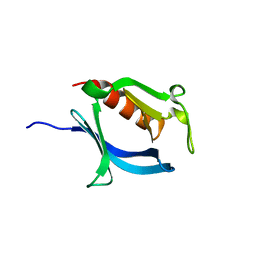 | |
3AJ4
 
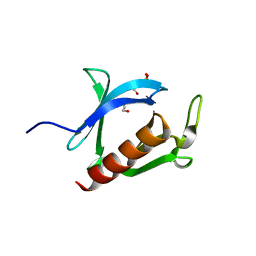 | | Crystal structure of the PH domain of Evectin-2 from human complexed with O-phospho-L-serine | | Descriptor: | 1,2-ETHANEDIOL, PHOSPHOSERINE, Pleckstrin homology domain-containing family B member 2 | | Authors: | Okazaki, S, Kato, R, Wakatsuki, S. | | Deposit date: | 2010-05-21 | | Release date: | 2011-05-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Intracellular phosphatidylserine is essential for retrograde membrane traffic through endosomes
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
8GS2
 
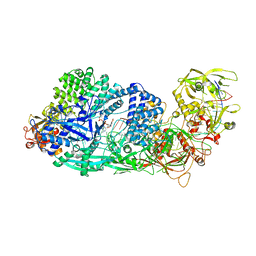 | | Structure of the Cas7-11-Csx29-guide RNA-target RNA (non-matching PFS) complex | | Descriptor: | ADENOSINE-5'-MONOPHOSPHATE, CHAT domain-containing protein, CRISPR-associated RAMP family protein, ... | | Authors: | Kato, K, Okazaki, S, Ishikawa, J, Isayama, Y, Nishizawa, T, Nishimasu, H. | | Deposit date: | 2022-09-04 | | Release date: | 2022-11-09 | | Last modified: | 2022-12-07 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | RNA-triggered protein cleavage and cell growth arrest by the type III-E CRISPR nuclease-protease.
Science, 378, 2022
|
|
4Y7P
 
 | | Structure of alkaline D-peptidase from Bacillus cereus | | Descriptor: | Alkaline D-peptidase, THIOCYANATE ION | | Authors: | Nakano, S, Okazaki, S, Ishitsubo, E, Kawahara, N, Komeda, H, Tokiwa, H, Asano, Y. | | Deposit date: | 2015-02-15 | | Release date: | 2015-10-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and computational analysis of peptide recognition mechanism of class-C type penicillin binding protein, alkaline D-peptidase from Bacillus cereus DF4-B
Sci Rep, 5, 2015
|
|
7CT4
 
 | | Crystal structure of D-amino acid oxidase from Rasamsonia emersonii strain YA | | Descriptor: | D-amino acid oxidase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Shimekake, Y, Hirato, Y, Okazaki, S, Funabashi, R, Goto, M, Furuichi, T, Suzuki, H, Takahashi, S. | | Deposit date: | 2020-08-18 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structure analysis of a unique D-amino-acid oxidase from the thermophilic fungus Rasamsonia emersonii strain YA.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
3HJE
 
 | | Crystal structure of sulfolobus tokodaii hypothetical maltooligosyl trehalose synthase | | Descriptor: | 704aa long hypothetical glycosyltransferase, GLYCEROL | | Authors: | Cielo, C.B.C, Okazaki, S, Suzuki, A, Mizushima, T, Masui, R, Kuramitsu, S, Yamane, T. | | Deposit date: | 2009-05-21 | | Release date: | 2010-04-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of ST0929, a putative glycosyl transferase from Sulfolobus tokodaii
Acta Crystallogr.,Sect.F, 66, 2010
|
|
7C7L
 
 | | Cryo-EM structure of the Cas12f1-sgRNA-target DNA complex | | Descriptor: | CRISPR-associated protein Cas14a.1, DNA (40-mer), ZINC ION, ... | | Authors: | Takeda, N.S, Nakagawa, R, Okazaki, S, Hirano, H, Kobayashi, K, Kusakizako, T, Nishizawa, T, Yamashita, K, Nishimasu, H, Nureki, O. | | Deposit date: | 2020-05-26 | | Release date: | 2020-12-23 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of the miniature type V-F CRISPR-Cas effector enzyme.
Mol.Cell, 81, 2021
|
|
7Y9Y
 
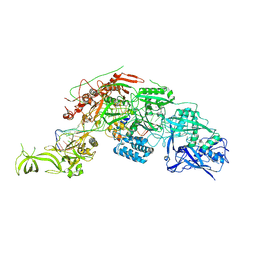 | | Structure of the Cas7-11-Csx29-guide RNA-target RNA (no PFS) complex | | Descriptor: | CHAT domain-containing protein, CRISPR-associated RAMP family protein, RNA (27-MER), ... | | Authors: | Kato, K, Okazaki, S, Ishikawa, J, Isayama, Y, Nishizawa, T, Nishimasu, H. | | Deposit date: | 2022-06-26 | | Release date: | 2022-11-09 | | Last modified: | 2022-12-07 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | RNA-triggered protein cleavage and cell growth arrest by the type III-E CRISPR nuclease-protease.
Science, 378, 2022
|
|
7Y9X
 
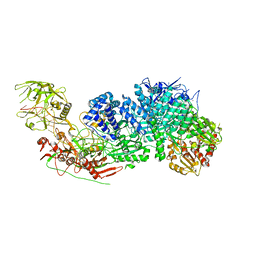 | | Structure of the Cas7-11-Csx29-guide RNA complex | | Descriptor: | CHAT domain-containing protein, CRISPR-associated RAMP family protein, ZINC ION, ... | | Authors: | Kato, K, Okazaki, S, Ishikawa, J, Isayama, Y, Nishizawa, T, Nishimasu, H. | | Deposit date: | 2022-06-26 | | Release date: | 2022-11-09 | | Last modified: | 2022-12-07 | | Method: | ELECTRON MICROSCOPY (2.49 Å) | | Cite: | RNA-triggered protein cleavage and cell growth arrest by the type III-E CRISPR nuclease-protease.
Science, 378, 2022
|
|
7WAH
 
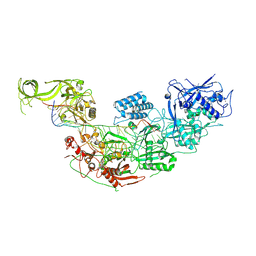 | | Structure of Cas7-11 in complex with guide RNA and target RNA | | Descriptor: | CRISPR-associated RAMP family protein, ZINC ION, crRNA (39-MER), ... | | Authors: | Kato, K, Okazaki, S, Isayama, Y, Nishizawa, T, Nishimasu, H. | | Deposit date: | 2021-12-14 | | Release date: | 2022-06-15 | | Last modified: | 2022-07-06 | | Method: | ELECTRON MICROSCOPY (2.45 Å) | | Cite: | Structure and engineering of the type III-E CRISPR-Cas7-11 effector complex.
Cell, 185, 2022
|
|
7YBD
 
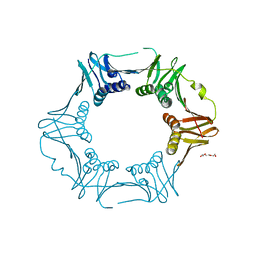 | | Crystal structure of sliding DNA clamp of Clostridioides difficile | | Descriptor: | Beta sliding clamp, TRIETHYLENE GLYCOL | | Authors: | Hishiki, A, Okazaki, S, Hara, K, Hashimoto, H. | | Deposit date: | 2022-06-29 | | Release date: | 2022-10-19 | | Last modified: | 2023-01-11 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Crystal structure of the sliding DNA clamp from the Gram-positive anaerobic bacterium Clostridioides difficile.
J.Biochem., 173, 2022
|
|
2EB9
 
 | | Crystal Structure of Cu(II)(Sal-Leu)/apo-Myoglobin | | Descriptor: | (N-SALICYLIDEN-L-LEUCINATO)-COPPER(II), GLYCEROL, Myoglobin, ... | | Authors: | Abe, S, Okazaki, S, Ueno, T, Hikage, T, Suzuki, A, Yamane, T, Watanabe, Y. | | Deposit date: | 2007-02-07 | | Release date: | 2007-07-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Design and Structure Analysis of Artificial Metalloproteins: Selective Coordination of His64 to Copper Complexes with Square-Planar Structure in the apo-Myoglobin Scaffold
Inorg.Chem., 46, 2007
|
|
2EB8
 
 | | Crystal Structure of Cu(II)(Sal-Phe)/apo-Myoglobin | | Descriptor: | (N-SALICYLIDEN-L-PHENYLALANATO)-COPPER(II), Myoglobin, PHOSPHATE ION | | Authors: | Abe, S, Okazaki, S, Ueno, T, Hikage, T, Suzuki, A, Yamane, T, Watanabe, Y. | | Deposit date: | 2007-02-07 | | Release date: | 2007-07-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Design and Structure Analysis of Artificial Metalloproteins: Selective Coordination of His64 to Copper Complexes with Square-Planar Structure in the apo-Myoglobin Scaffold
Inorg.Chem., 46, 2007
|
|
2EF2
 
 | | Crystal Structure of an Artificial Metalloprotein:Rh(Phebox-Ph)/apo-A71G Myoglobin | | Descriptor: | Myoglobin, PHOSPHATE ION, [2,6-BIS(4-PHENYL)-1,3-OXAZOLIN-2-YL]RHODIUM(III) | | Authors: | Abe, S, Satake, Y, Okazaki, S, Ueno, T, Hikage, T, Suzuki, A, Yamane, T, Nakajima, H, Watanabe, Y. | | Deposit date: | 2007-02-20 | | Release date: | 2007-10-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Incorporation of a Phebox Rhodium Complex into apo-Myoglobin Affords a Stable Organometallic Protein Showing Unprecedented Arrangement of the Complex in the Cavity
ORGANOMETALLICS, 26, 2007
|
|
5ZBC
 
 | | Crystal structure of Se-Met tryptophan oxidase (C395A mutant) from Chromobacterium violaceum | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Flavin-dependent L-tryptophan oxidase VioA | | Authors: | Yamaguchi, H, Tatsumi, M, Takahashi, K, Tagami, U, Sugiki, M, Kashiwagi, T, Okazaki, S, Mizukoshi, T, Asano, Y. | | Deposit date: | 2018-02-11 | | Release date: | 2018-12-19 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Protein engineering for improving the thermostability of tryptophan oxidase and insights from structural analysis.
J. Biochem., 164, 2018
|
|
