1GZ0
 
 | | 23S RIBOSOMAL RNA G2251 2'O-METHYLTRANSFERASE RLMB | | Descriptor: | HYPOTHETICAL TRNA/RRNA METHYLTRANSFERASE YJFH | | Authors: | Michel, G, Cygler, M. | | Deposit date: | 2002-05-03 | | Release date: | 2002-10-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Structure of the Rlmb 23S Rrna Methyltransferase Reveals a New Methyltransferase Fold with a Unique Knot
Structure, 10, 2002
|
|
1KTW
 
 | | IOTA-CARRAGEENASE COMPLEXED TO IOTA-CARRAGEENAN FRAGMENTS | | Descriptor: | 3,6-anhydro-2-O-sulfo-alpha-D-galactopyranose-(1-3)-4-O-sulfo-beta-D-galactopyranose, 3,6-anhydro-2-O-sulfo-alpha-D-galactopyranose-(1-3)-4-O-sulfo-beta-D-galactopyranose-(1-4)-3,6-anhydro-2-O-sulfo-alpha-D-galactopyranose-(1-3)-4-O-sulfo-beta-D-galactopyranose, CALCIUM ION, ... | | Authors: | Michel, G, Kahn, R, Dideberg, O. | | Deposit date: | 2002-01-18 | | Release date: | 2003-06-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Structural Bases of the Processive Degradation of iota-Carrageenan, a Main Cell Wall Polysaccharide of Red Algae.
J.Mol.Biol., 334, 2003
|
|
1DYP
 
 | | 1,3-ALPHA-1,4-BETA-D-GALACTOSE-4-SULFATE-3,6-ANHYDRO-D-GALACTOSE 4 GALACTOHYDROLASE | | Descriptor: | CADMIUM ION, CHLORIDE ION, KAPPA-CARRAGEENASE | | Authors: | Michel, G, Chantalat, L, Dideberg, O. | | Deposit date: | 2000-02-04 | | Release date: | 2001-01-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | The Kappa-Carrageenase of P. Carrageenovora Features a Tunnel-Shaped Active Site: A Novel Insight in the Evolution of Clan-B Glycoside Hydrolases
Structure, 9, 2001
|
|
1H80
 
 | | 1,3-ALPHA-1,4-BETA-D-GALACTOSE-4-SULFATE- 3,6-ANHYDRO-D-GALACTOSE-2-SULFATE 4 GALACTOHYDROLASE | | Descriptor: | CALCIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Michel, G, Chantalat, L, Dideberg, O. | | Deposit date: | 2001-01-22 | | Release date: | 2001-11-27 | | Last modified: | 2018-10-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Iota-Carrageenase of Alteromonas Fortis. A Beta-Helix Fold-Containing Enzyme for the Degradation of a Highly Polyanionic Polysaccharide
J.Biol.Chem., 276, 2001
|
|
1O9B
 
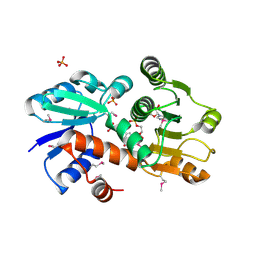 | | QUINATE/SHIKIMATE DEHYDROGENASE YDIB COMPLEXED WITH NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, HYPOTHETICAL SHIKIMATE 5-DEHYDROGENASE-LIKE PROTEIN YDIB, PHOSPHATE ION | | Authors: | Michel, G, Cygler, M. | | Deposit date: | 2002-12-12 | | Release date: | 2003-02-01 | | Last modified: | 2013-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of Shikimate Dehydrogenase Aroe and its Paralog Ydib: A Common Structural Framework for Different Activities
J.Biol.Chem., 278, 2003
|
|
1OFL
 
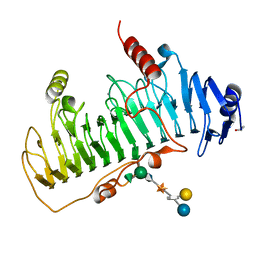 | | CRYSTAL STRUCTURE OF CHONDROITINASE B COMPLEXED TO DERMATAN SULFATE HEXASACCHARIDE | | Descriptor: | 4-deoxy-alpha-L-threo-hex-4-enopyranuronic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-alpha-D-galactopyranose, 4-deoxy-alpha-L-threo-hex-4-enopyranuronic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose, CALCIUM ION, ... | | Authors: | Michel, G, Cygler, M. | | Deposit date: | 2003-04-15 | | Release date: | 2004-06-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Structure of Chondroitin B Lyase Complexed with Glycosaminoglycan Oligosaccharides Unravels a Calcium-Dependent Catalytic Machinery
J.Biol.Chem., 279, 2004
|
|
1OFM
 
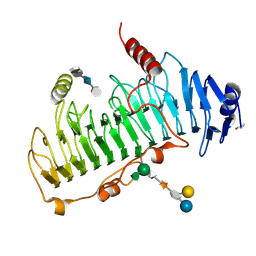 | | CRYSTAL STRUCTURE OF CHONDROITINASE B COMPLEXED TO CHONDROITIN 4-SULFATE TETRASACCHARIDE | | Descriptor: | 4-deoxy-alpha-L-threo-hex-4-enopyranuronic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose, CHONDROITINASE B, alpha-D-galactopyranose-(1-3)-[beta-D-glucopyranose-(1-4)]2-O-methyl-alpha-L-fucopyranose-(1-4)-beta-D-xylopyranose-(1-4)-alpha-D-glucopyranuronic acid-(1-2)-[alpha-L-rhamnopyranose-(1-4)]alpha-D-mannopyranose | | Authors: | Michel, G, Cygler, M. | | Deposit date: | 2003-04-15 | | Release date: | 2004-04-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Structure of Chondroitin B Lyase Complexed with Glycosaminoglycan Oligosaccharides Unravels a Calcium-Dependent Catalytic Machinery
J.Biol.Chem., 279, 2004
|
|
5OLC
 
 | |
5FUI
 
 | | Crystal structure of the C-terminal CBM6 of LamC a marine laminarianse from Zobellia galactanivorans | | Descriptor: | 2-AMINOMETHYL-PYRIDINE, ENDO-1,3-BETA-GLUCANASE, FAMILY GH16, ... | | Authors: | Labourel, A, Jam, M, Legentil, L, Sylla, B, Hehemann, J.H, Ficko-Blean, E, Ferrieres, V, Czjzek, M, Michel, G. | | Deposit date: | 2016-01-27 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Unraveling the Multivalent Binding of a Marine Family 6 Carbohydrate-Binding Module with its Native Laminarin Ligand.
FEBS J., 283, 2016
|
|
4USZ
 
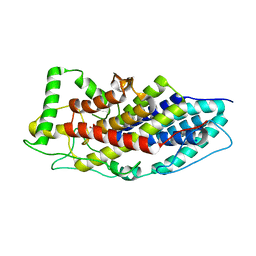 | | Crystal structure of the first bacterial vanadium dependant iodoperoxidase | | Descriptor: | SODIUM ION, VANADATE ION, VANADIUM-DEPENDENT HALOPEROXIDASE | | Authors: | Rebuffet, E, Delage, L, Fournier, J.B, Rzonca, J, Potin, P, Michel, G, Czjzek, M, Leblanc, C. | | Deposit date: | 2014-07-17 | | Release date: | 2014-10-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Bacterial Vanadium Iodoperoxidase from the Marine Flavobacteriaceae Zobellia Galactanivorans Reveals Novel Molecular and Evolutionary Features of Halide Specificity in This Enzyme Family.
Appl.Environ.Microbiol., 80, 2014
|
|
4CRQ
 
 | | Crystal structure of the catalytic domain of the modular laminarinase ZgLamC mutant E142S | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Labourel, A, Jam, M, Legentil, L, Sylla, B, Ficko-Blean, E, Hehemann, J.H, Ferrieres, V, Czjzek, M, Michel, G. | | Deposit date: | 2014-02-28 | | Release date: | 2015-01-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and Biochemical Characterization of the Laminarina Zglamc[Gh16] from Zobellia Galactanivorans Suggests Preferred Recognition of Branched Laminarin
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3LMW
 
 | | Crystal structure of iota-carrageenase family GH82 from A. fortis in absence of chloride ions | | Descriptor: | CALCIUM ION, Iota-carrageenase, CgiA, ... | | Authors: | Rebuffet, E, Barbeyron, T, Jeudy, A, Czjzek, M, Michel, G. | | Deposit date: | 2010-02-01 | | Release date: | 2010-08-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Identification of catalytic residues and mechanistic analysis of family GH82 iota-carrageenases
Biochemistry, 49, 2010
|
|
2UWB
 
 | | Crystal structure of the Nasturtium seedling mutant xyloglucanase isoform NXG1-delta-YNIIG | | Descriptor: | CELLULASE | | Authors: | Baumann, M.J, Eklof, J, Michel, G, Kallasa, A, Teeri, T.T, Brumer, H, Czjzek, M. | | Deposit date: | 2007-03-20 | | Release date: | 2007-06-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Evidence for the Evolution of Xyloglucanase Activity from Xyloglucan Endo-Transglycosylases: Biological Implications for Cell Wall Metabolism.
Plant Cell, 19, 2007
|
|
2UWA
 
 | | Crystal structure of the Nasturtium seedling xyloglucanase isoform NXG1 | | Descriptor: | CELLULASE, GLYCEROL | | Authors: | Baumann, M.J, Eklof, J, Michel, G, Kallasa, A, Teeri, T.T, Brumer, H, Czjzek, M. | | Deposit date: | 2007-03-19 | | Release date: | 2007-06-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Evidence for the Evolution of Xyloglucanase Activity from Xyloglucan Endo-Transglycosylases: Biological Implications for Cell Wall Metabolism.
Plant Cell, 19, 2007
|
|
2UWC
 
 | | Crystal structure of Nasturtium xyloglucan hydrolase isoform NXG2 | | Descriptor: | CELLULASE | | Authors: | Baumann, M.J, Eklof, J.M, Michel, G, Kallas, A, Teeri, T.T, Brumer, H, Czjzek, M. | | Deposit date: | 2007-03-20 | | Release date: | 2007-06-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Evidence for the Evolution of Xyloglucanase Activity from Xyloglucan Endo-Transglycosylases: Biological Implications for Cell Wall Metabolism.
Plant Cell, 19, 2007
|
|
2VH9
 
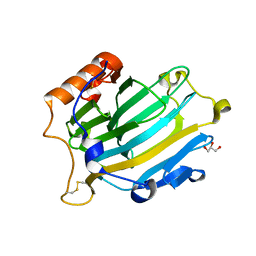 | | CRYSTAL STRUCTURE OF NXG1-DELTAYNIIG IN COMPLEX WITH XLLG, A XYLOGLUCAN DERIVED OLIGOSACCHARIDE | | Descriptor: | CELLULASE, GLYCEROL, ZINC ION, ... | | Authors: | Czjzek, M, Mark, P, Baumann, M.J, Eklof, J.M, Michel, G, Brumer, H. | | Deposit date: | 2007-11-20 | | Release date: | 2008-11-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Analysis of Nasturtium Tmnxg1 Complexes by Crystallography and Molecular Dynamics Provides Detailed Insight Into Substrate Recognition by Family Gh16 Xyloglucan Endo-Transglycosylases and Endo-Hydrolases.
Proteins, 75, 2009
|
|
4ATE
 
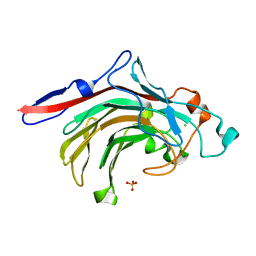 | | High resolution crystal structure of beta-porphyranase A from Zobellia galactanivorans | | Descriptor: | BETA-PORPHYRANASE A, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Hehemann, J.H, Correc, G, Jam, M, Michel, G, Czjzek, M. | | Deposit date: | 2012-05-06 | | Release date: | 2012-07-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Biochemical and Structural Characterization of the Complex Agarolytic Enzyme System from the Marine Bacterium Zobellia Galactanivorans.
J.Biol.Chem., 287, 2012
|
|
4ATF
 
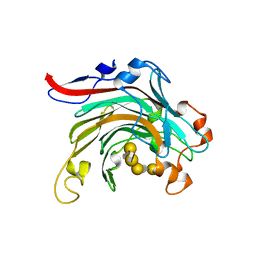 | | Crystal structure of inactivated mutant beta-agarase B in complex with agaro-octaose | | Descriptor: | 3,6-anhydro-alpha-L-galactopyranose-(1-3)-beta-D-galactopyranose-(1-4)-3,6-anhydro-alpha-L-galactopyranose-(1-3)-beta-D-galactopyranose-(1-4)-3,6-anhydro-alpha-L-galactopyranose-(1-3)-beta-D-galactopyranose-(1-4)-3,6-anhydro-alpha-L-galactopyranose-(1-3)-beta-D-galactopyranose, BETA-AGARASE B, SODIUM ION | | Authors: | Bernard, T, Hehemann, J.H, Correc, G, Jam, M, Michel, G, Czjzek, M. | | Deposit date: | 2012-05-06 | | Release date: | 2012-07-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Biochemical and Structural Characterization of the Complex Agarolytic Enzyme System from the Marine Bacterium Zobellia Galactanivorans.
J.Biol.Chem., 287, 2012
|
|
3ILF
 
 | | Crystal structure of porphyranase A (PorA) in complex with neo-porphyrotetraose | | Descriptor: | 6-O-sulfo-alpha-L-galactopyranose-(1-3)-beta-D-galactopyranose-(1-4)-6-O-sulfo-alpha-L-galactopyranose-(1-3)-alpha-D-galactopyranose, ACETATE ION, CALCIUM ION, ... | | Authors: | Hehemann, J.H, Correc, G, Barbeyron, T, Helbert, W, Michel, G, Czjzek, M. | | Deposit date: | 2009-08-07 | | Release date: | 2010-04-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Transfer of carbohydrate-active enzymes from marine bacteria to Japanese gut microbiota.
Nature, 464, 2010
|
|
3JUU
 
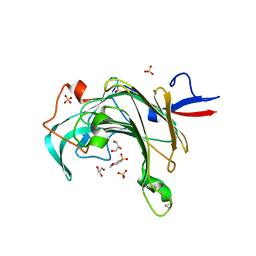 | | Crystal structure of porphyranase B (PorB) from Zobellia galactanivorans | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, SULFATE ION, ... | | Authors: | Hehemann, J.H, Correc, G, Barbeyron, T, Michel, G, Czjzek, M. | | Deposit date: | 2009-09-15 | | Release date: | 2010-04-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Transfer of carbohydrate-active enzymes from marine bacteria to Japanese gut microbiota.
Nature, 464, 2010
|
|
3P2N
 
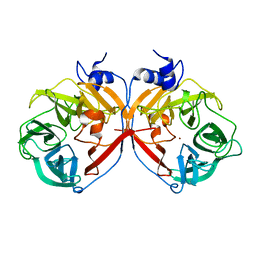 | | Discovery and structural characterization of a new glycoside hydrolase family abundant in coastal waters that was annotated as 'hypothetical protein' | | Descriptor: | 3,6-anhydro-alpha-L-galactosidase, CHLORIDE ION, ZINC ION | | Authors: | Rebuffet, E, Barbeyron, T, Czjzek, M, Michel, G. | | Deposit date: | 2010-10-03 | | Release date: | 2011-03-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Discovery and structural characterization of a novel glycosidase family of marine origin.
Environ Microbiol, 13, 2011
|
|
4ASM
 
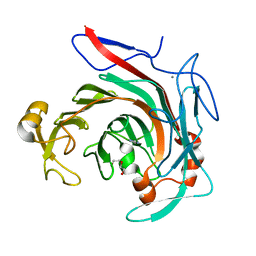 | | Crystal structure of the catalytic domain of beta-agarase D from Zobellia galactanivorans | | Descriptor: | BETA-AGARASE D, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Hehemann, J.H, Correc, G, Bernard, T, Michel, G, Czjzek, M. | | Deposit date: | 2012-05-02 | | Release date: | 2012-07-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Biochemical and Structural Characterization of the Complex Agarolytic Enzyme System from the Marine Bacterium Zobellia Galactanivorans.
J.Biol.Chem., 287, 2012
|
|
5OPQ
 
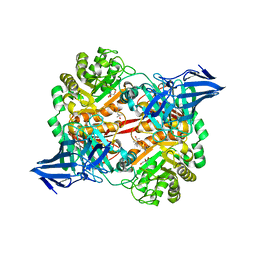 | | A 3,6-anhydro-D-galactosidase produced by Zobellia galactanivorans. This is an exo-lytic enzyme that hydrolyzes terminal 3,6-anhydro-D-galactose from the non-reducing end of carrageenan oligosaccharides. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3,6-anhydro-D-galactosidase, ... | | Authors: | Ficko-Blean, E, Michel, G, Czjzek, M. | | Deposit date: | 2017-08-10 | | Release date: | 2017-11-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Carrageenan catabolism is encoded by a complex regulon in marine heterotrophic bacteria.
Nat Commun, 8, 2017
|
|
4BE3
 
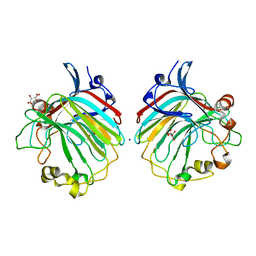 | | crystal structure of the exolytic PL7 alginate lyase AlyA5 from Zobellia galactanivorans | | Descriptor: | ALGINATE LYASE, FAMILY PL7, D(-)-TARTARIC ACID, ... | | Authors: | Thomas, F, Jeudy, A, Michel, G, Czjzek, M. | | Deposit date: | 2013-03-05 | | Release date: | 2013-06-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Comparative Characterization of Two Marine Alginate Lyases from Zobellia Galactanivorans Reveals Distinct Modes of Action and Exquisite Adaptation to Their Natural Substrate.
J.Biol.Chem., 288, 2013
|
|
3ZPY
 
 | | Crystal structure of the marine PL7 alginate lyase AlyA1 from Zobellia galactanivorans | | Descriptor: | ALGINATE LYASE, FAMILY PL7, SODIUM ION | | Authors: | Thomas, F, Jeudy, A, Michel, G, Czjzek, M. | | Deposit date: | 2013-03-04 | | Release date: | 2013-06-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Comparative Characterization of Two Marine Alginate Lyases from Zobellia Galactanivorans Reveals Distinct Modes of Action and Exquisite Adaptation to Their Natural Substrate.
J.Biol.Chem., 288, 2013
|
|
