2ADL
 
 | | Solution structure of the bacterial antitoxin CcdA: Implications for DNA and toxin binding | | Descriptor: | CcdA | | Authors: | Madl, T, VanMelderen, L, Oberer, M, Keller, W, Khatai, L, Zangger, K. | | Deposit date: | 2005-07-20 | | Release date: | 2006-08-22 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural basis for nucleic acid and toxin recognition of the bacterial antitoxin CcdA
J.Mol.Biol., 364, 2006
|
|
2ADN
 
 | | Solution structure of the bacterial antitoxin CcdA: Implications for DNA and toxin binding | | Descriptor: | CcdA | | Authors: | Madl, T, VanMelderen, L, Oberer, M, Keller, W, Khatai, L, Zangger, K. | | Deposit date: | 2005-07-20 | | Release date: | 2006-08-22 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural basis for nucleic acid and toxin recognition of the bacterial antitoxin CcdA
J.Mol.Biol., 364, 2006
|
|
2KLF
 
 | | PERE NMR structure of maltodextrin-binding protein | | Descriptor: | Maltose-binding periplasmic protein | | Authors: | Madl, T, Bermel, W, Zangger, K. | | Deposit date: | 2009-07-02 | | Release date: | 2009-10-06 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Use of Relaxation Enhancements in a Paramagnetic Environment for the Structure Determination of Proteins Using NMR Spectroscopy
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
2KLG
 
 | | PERE NMR structure of ubiquitin | | Descriptor: | Ubiquitin | | Authors: | Madl, T, Bermel, W, Zangger, K. | | Deposit date: | 2009-07-02 | | Release date: | 2009-10-06 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Use of Relaxation Enhancements in a Paramagnetic Environment for the Structure Determination of Proteins Using NMR Spectroscopy
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
2H3A
 
 | | Structural basis for nucleic acid and toxin recognition of the bacterial antitoxin CcdA | | Descriptor: | 5'-D(P*AP*TP*AP*TP*GP*TP*AP*TP*AP*CP*CP*CP*G)-3', 5'-D(P*TP*CP*GP*GP*GP*TP*AP*TP*AP*CP*AP*TP*A)-3', CcdA | | Authors: | Madl, T, Van Melderen, L, Respondek, M, Oberer, M, Keller, W, Zangger, K. | | Deposit date: | 2006-05-22 | | Release date: | 2006-11-21 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for Nucleic Acid and Toxin Recognition of the Bacterial Antitoxin CcdA
J.Mol.Biol., 364, 2006
|
|
2H3C
 
 | | Structural basis for nucleic acid and toxin recognition of the bacterial antitoxin CcdA | | Descriptor: | 5'-D(P*AP*TP*AP*TP*GP*TP*AP*TP*AP*CP*CP*CP*G)-3', 5'-D(P*TP*CP*GP*GP*GP*TP*AP*TP*AP*CP*AP*TP*A)-3', CcdA | | Authors: | Madl, T, Van Melderen, L, Respondek, M, Oberer, M, Keller, W, Zangger, K. | | Deposit date: | 2006-05-22 | | Release date: | 2006-11-21 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for Nucleic Acid and Toxin Recognition of the Bacterial Antitoxin CcdA
J.Mol.Biol., 364, 2006
|
|
2L1L
 
 | |
2M0G
 
 | | Structure, phosphorylation and U2AF65 binding of the Nterminal Domain of splicing factor 1 during 3 splice site Recognition | | Descriptor: | Splicing factor 1, Splicing factor U2AF 65 kDa subunit | | Authors: | Madl, T, Sattler, M, Zhang, Y, Bagdiul, I, Kern, T, Kang, H, Zou, P, Maeusbacher, N, Sieber, S.A, Kraemer, A. | | Deposit date: | 2012-10-25 | | Release date: | 2013-01-30 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure, phosphorylation and U2AF65 binding of the N-terminal domain of splicing factor 1 during 3'-splice site recognition.
Nucleic Acids Res., 41, 2013
|
|
2M09
 
 | | Structure, phosphorylation and U2AF65 binding of the Nterminal Domain of splicing factor 1 during 3 splice site Recognition | | Descriptor: | Splicing factor 1 | | Authors: | Madl, T, Sattler, M, Zhang, Y, Bagdiul, I, Kern, T, Kang, H, Zou, P, Maeusbacher, N, Sieber, S.A, Kraemer, A. | | Deposit date: | 2012-10-22 | | Release date: | 2013-01-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure, phosphorylation and U2AF65 binding of the N-terminal domain of splicing factor 1 during 3'-splice site recognition.
Nucleic Acids Res., 41, 2013
|
|
8CMK
 
 | | Transportin-3 TNPO3 in complex with RSY region of CIRBP | | Descriptor: | 2-(1H-INDOL-3-YL)ETHANAMINE, Cold-inducible RNA-binding protein, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Zhou, Q, Sagmeister, T, Pavkov-Keller, T, Madl, T. | | Deposit date: | 2023-02-20 | | Release date: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.945 Å) | | Cite: | Tyrosine mediated nuclear import of CIRBP reveals a flexible NLS recognition by TNPO3
To Be Published
|
|
4CZ3
 
 | | HP24wt derived from the villin headpiece subdomain | | Descriptor: | VILLIN-1 | | Authors: | Hocking, H, Haese, F, Madl, T, Zacharias, M, Rief, M, Zoldak, G. | | Deposit date: | 2014-04-16 | | Release date: | 2015-02-18 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | A Compact Native 24-Residue Supersecondary Structure Derived from the Villin Headpiece Subdomain.
Biophys.J., 108, 2015
|
|
4CZ4
 
 | | HP24stab derived from the villin headpiece subdomain | | Descriptor: | VILLIN-1 | | Authors: | Hocking, H, Haese, F, Madl, T, Zacharias, M, Rief, M, Zoldak, G. | | Deposit date: | 2014-04-16 | | Release date: | 2015-02-18 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | A Compact Native 24-Residue Supersecondary Structure Derived from the Villin Headpiece Subdomain.
Biophys.J., 108, 2015
|
|
7NMB
 
 | |
4EL6
 
 | | Crystal structure of IPSE/alpha-1 from Schistosoma mansoni eggs | | Descriptor: | IL-4-inducing protein | | Authors: | Mayerhofer, H, Meyer, H, Tripsianes, K, Barths, D, Blindow, S, Bade, S, Madl, T, Frey, A, Haas, H, Sattler, M, Schramm, G, Mueller-Dieckmann, J. | | Deposit date: | 2012-04-10 | | Release date: | 2013-04-10 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Structure and functional analysis of IPSE/alpha-1, an IL-4-inducing factor secreted from Schistosoma mansoni eggs, reveals an IgE-binding crystallin fold
To be Published
|
|
4JVU
 
 | | IgM C2-domain from mouse | | Descriptor: | Ig mu chain C region membrane-bound form | | Authors: | Mueller, R, Graewert, A.M, Kern, T, Madl, T, Peschek, J, Sattler, M, Groll, M, Buchner, J. | | Deposit date: | 2013-03-26 | | Release date: | 2013-06-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | High-resolution structures of the IgM Fc domains reveal principles of its hexamer formation.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4JVW
 
 | | IgM C4-domain from mouse | | Descriptor: | Ig mu chain C region secreted form | | Authors: | Mueller, R, Graewert, A.M, Kern, T, Madl, T, Peschek, J, Sattler, M, Groll, M, Buchner, J. | | Deposit date: | 2013-03-26 | | Release date: | 2013-06-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High-resolution structures of the IgM Fc domains reveal principles of its hexamer formation.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3NBY
 
 | | Crystal structure of the PKI NES-CRM1-RanGTP nuclear export complex | | Descriptor: | Exportin-1, GTP-binding nuclear protein Ran, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Guttler, T, Madl, T, Neumann, P, Deichsel, D, Corsini, L, Monecke, T, Ficner, R, Sattler, M, Gorlich, D. | | Deposit date: | 2010-06-04 | | Release date: | 2010-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.42 Å) | | Cite: | NES consensus redefined by structures of PKI-type and Rev-type nuclear export signals bound to CRM1.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3NC0
 
 | | Crystal structure of the HIV-1 Rev NES-CRM1-RanGTP nuclear export complex (crystal II) | | Descriptor: | DI(HYDROXYETHYL)ETHER, Exportin-1, GLYCEROL, ... | | Authors: | Guttler, T, Madl, T, Neumann, P, Deichsel, D, Corsini, L, Monecke, T, Ficner, R, Sattler, M, Gorlich, D. | | Deposit date: | 2010-06-04 | | Release date: | 2010-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | NES consensus redefined by structures of PKI-type and Rev-type nuclear export signals bound to CRM1.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3NBZ
 
 | | Crystal structure of the HIV-1 Rev NES-CRM1-RanGTP nuclear export complex (crystal I) | | Descriptor: | Exportin-1, GTP-binding nuclear protein Ran, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Guttler, T, Madl, T, Neumann, P, Deichsel, D, Corsini, L, Monecke, T, Ficner, R, Sattler, M, Gorlich, D. | | Deposit date: | 2010-06-04 | | Release date: | 2010-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | NES consensus redefined by structures of PKI-type and Rev-type nuclear export signals bound to CRM1.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3NC1
 
 | | Crystal structure of the CRM1-RanGTP complex | | Descriptor: | Exportin-1, GTP-binding nuclear protein Ran, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Guttler, T, Madl, T, Neumann, P, Deichsel, D, Corsini, L, Monecke, T, Ficner, R, Sattler, M, Gorlich, D. | | Deposit date: | 2010-06-04 | | Release date: | 2010-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | NES consensus redefined by structures of PKI-type and Rev-type nuclear export signals bound to CRM1.
Nat.Struct.Mol.Biol., 17, 2010
|
|
2YH1
 
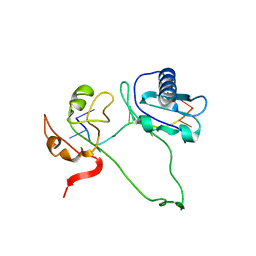 | | Model of human U2AF65 tandem RRM1 and RRM2 domains with eight-site uridine binding | | Descriptor: | 5'-R(*UP*UP*UP*UP*UP*UP*UP*UP*UP)-3', SPLICING FACTOR U2AF 65 KDA SUBUNIT | | Authors: | Mackereth, C.D, Madl, T, Simon, B, Zanier, K, Gasch, A, Sattler, M. | | Deposit date: | 2011-04-26 | | Release date: | 2011-07-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Multi-Domain Conformational Selection Underlies Pre-Mrna Splicing Regulation by U2Af
Nature, 475, 2011
|
|
2W84
 
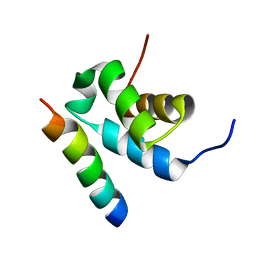 | | Structure of Pex14 in complex with Pex5 | | Descriptor: | PEROXISOMAL MEMBRANE PROTEIN PEX14, PEROXISOMAL TARGETING SIGNAL 1 RECEPTOR | | Authors: | Neufeld, C, Filipp, F.V, Simon, B, Neuhaus, A, Schueller, N, David, C, Kooshapur, H, Madl, T, Erdmann, R, Schliebs, W, Wilmanns, M, Sattler, M. | | Deposit date: | 2009-01-09 | | Release date: | 2009-02-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for competitive interactions of Pex14 with the import receptors Pex5 and Pex19.
EMBO J., 28, 2009
|
|
2W85
 
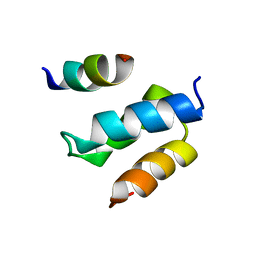 | | Structure of Pex14 in complex with Pex19 | | Descriptor: | PEROXIN-19, PEROXISOMAL MEMBRANE ANCHOR PROTEIN PEX14 | | Authors: | Neufeld, C, Filipp, F.V, Simon, B, Neuhaus, A, Schueller, N, David, C, Kooshapur, H, Madl, T, Erdmann, R, Schliebs, W, Wilmanns, M, Sattler, M. | | Deposit date: | 2009-01-09 | | Release date: | 2009-02-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for Competitive Interactions of Pex14 with the Import Receptors Pex5 and Pex19.
Embo J., 28, 2009
|
|
2YH0
 
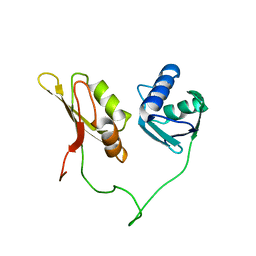 | | Solution structure of the closed conformation of human U2AF65 tandem RRM1 and RRM2 domains | | Descriptor: | SPLICING FACTOR U2AF 65 KDA SUBUNIT | | Authors: | Mackereth, C.D, Madl, T, Simon, B, Zanier, K, Gasch, A, Sattler, M. | | Deposit date: | 2011-04-26 | | Release date: | 2011-07-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Multi-Domain Conformational Selection Underlies Pre-Mrna Splicing Regulation by U2Af
Nature, 475, 2011
|
|
2XA6
 
 | | Structural basis for homodimerization of the Src-associated during mitosis, 68 kD protein (Sam68) Qua1 domain | | Descriptor: | KH DOMAIN-CONTAINING,RNA-BINDING,SIGNAL TRANSDUCTION-ASSOCIATED PROTEIN 1 | | Authors: | Meyer, N.H, Tripsianes, K, Vincendeaux, M, Madl, T, Kateb, F, Brack-Werner, R, Sattler, M. | | Deposit date: | 2010-03-29 | | Release date: | 2010-07-07 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structural basis for homodimerization of the Src-associated during mitosis, 68-kDa protein (Sam68) Qua1 domain.
J. Biol. Chem., 285, 2010
|
|
