1HSA
 
 | | THE THREE-DIMENSIONAL STRUCTURE OF HLA-B27 AT 2.1 ANGSTROMS RESOLUTION SUGGESTS A GENERAL MECHANISM FOR TIGHT PEPTIDE BINDING TO MHC | | Descriptor: | BETA 2-MICROGLOBULIN, CLASS I HISTOCOMPATIBILITY ANTIGEN (HLA-B*2705), MODEL PEPTIDE SEQUENCE - ARAAAAAAA | | Authors: | Madden, D.R, Gorga, J.C, Strominger, J.L, Wiley, D.C. | | Deposit date: | 1992-08-11 | | Release date: | 1992-10-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The three-dimensional structure of HLA-B27 at 2.1 A resolution suggests a general mechanism for tight peptide binding to MHC.
Cell(Cambridge,Mass.), 70, 1992
|
|
1HHG
 
 | | THE ANTIGENIC IDENTITY OF PEPTIDE(SLASH)MHC COMPLEXES: A COMPARISON OF THE CONFORMATION OF FIVE PEPTIDES PRESENTED BY HLA-A2 | | Descriptor: | BETA 2-MICROGLOBULIN, CLASS I HISTOCOMPATIBILITY ANTIGEN (HLA-A*0201) (ALPHA CHAIN), HIV-1 GP120 ENVELOPE PROTEIN (RESIDUES 195-207) | | Authors: | Madden, D.R, Garboczi, D.N, Wiley, D.C. | | Deposit date: | 1993-06-30 | | Release date: | 1993-10-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The antigenic identity of peptide-MHC complexes: a comparison of the conformations of five viral peptides presented by HLA-A2.
Cell(Cambridge,Mass.), 75, 1993
|
|
1HHH
 
 | | THE ANTIGENIC IDENTITY OF PEPTIDE(SLASH)MHC COMPLEXES: A COMPARISON OF THE CONFORMATION OF FIVE PEPTIDES PRESENTED BY HLA-A2 | | Descriptor: | BETA 2-MICROGLOBULIN, CLASS I HISTOCOMPATIBILITY ANTIGEN (HLA-A*0201) (ALPHA CHAIN), HEPATITIS B NUCLEOCAPSID PROTEIN (RESIDUES 18-27) | | Authors: | Madden, D.R, Garboczi, D.N, Wiley, D.C. | | Deposit date: | 1993-06-30 | | Release date: | 1993-10-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The antigenic identity of peptide-MHC complexes: a comparison of the conformations of five viral peptides presented by HLA-A2.
Cell(Cambridge,Mass.), 75, 1993
|
|
1HHK
 
 | | THE ANTIGENIC IDENTITY OF PEPTIDE(SLASH)MHC COMPLEXES: A COMPARISON OF THE CONFORMATION OF FIVE PEPTIDES PRESENTED BY HLA-A2 | | Descriptor: | BETA 2-MICROGLOBULIN, CLASS I HISTOCOMPATIBILITY ANTIGEN (HLA-A*0201) (ALPHA CHAIN), NONAMERIC PEPTIDE FROM HTLV-1 TAX PROTEIN (RESIDUES 11-19) | | Authors: | Madden, D.R, Garboczi, D.N, Wiley, D.C. | | Deposit date: | 1993-06-30 | | Release date: | 1993-10-31 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The antigenic identity of peptide-MHC complexes: a comparison of the conformations of five viral peptides presented by HLA-A2.
Cell(Cambridge,Mass.), 75, 1993
|
|
1HHJ
 
 | | THE ANTIGENIC IDENTITY OF PEPTIDE(SLASH)MHC COMPLEXES: A COMPARISON OF THE CONFORMATION OF FIVE PEPTIDES PRESENTED BY HLA-A2 | | Descriptor: | BETA 2-MICROGLOBULIN, CLASS I HISTOCOMPATIBILITY ANTIGEN (HLA-A*0201) (ALPHA CHAIN), HIV-1 REVERSE TRANSCRIPTASE (RESIDUES 309-317) | | Authors: | Madden, D.R, Garboczi, D.N, Wiley, D.C. | | Deposit date: | 1993-06-30 | | Release date: | 1993-10-31 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The antigenic identity of peptide-MHC complexes: a comparison of the conformations of five viral peptides presented by HLA-A2.
Cell(Cambridge,Mass.), 75, 1993
|
|
1HHI
 
 | | THE ANTIGENIC IDENTITY OF PEPTIDE(SLASH)MHC COMPLEXES: A COMPARISON OF THE CONFORMATION OF FIVE PEPTIDES PRESENTED BY HLA-A2 | | Descriptor: | BETA 2-MICROGLOBULIN, CLASS I HISTOCOMPATIBILITY ANTIGEN (HLA-A*0201) (ALPHA CHAIN), INFLUENZA A MATRIX PROTEIN M1 (RESIDUES 58-66) | | Authors: | Madden, D.R, Garboczi, D.N, Wiley, D.C. | | Deposit date: | 1993-06-30 | | Release date: | 1993-10-31 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The antigenic identity of peptide-MHC complexes: a comparison of the conformations of five viral peptides presented by HLA-A2.
Cell(Cambridge,Mass.), 75, 1993
|
|
2V9A
 
 | | Structure of Citrate-free Periplasmic Domain of Sensor Histidine Kinase CitA | | Descriptor: | SENSOR KINASE CITA | | Authors: | Sevvana, M, Vijayan, V, Zweckstetter, M, Reinelt, S, Madden, D.R, Sheldrick, G.M, Bott, M, Griesinger, C, Becker, S. | | Deposit date: | 2007-08-23 | | Release date: | 2008-03-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Ligand-Induced Switch in the Periplasmic Domain of Sensor Histidine Kinase Cita.
J.Mol.Biol., 377, 2008
|
|
8F6U
 
 | |
8F6V
 
 | |
8GJR
 
 | |
3SFJ
 
 | |
6OV7
 
 | | CFTR Associated Ligand (CAL) PDZ domain bound to peptide kCAL01 | | Descriptor: | Golgi-associated PDZ and coiled-coil motif-containing protein, kCAL01 peptide | | Authors: | Gill, N.P, Madden, D.R. | | Deposit date: | 2019-05-07 | | Release date: | 2019-11-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Computational Analysis of Energy Landscapes Reveals Dynamic Features That Contribute to Binding of Inhibitors to CFTR-Associated Ligand.
J.Phys.Chem.B, 123, 2019
|
|
4YX9
 
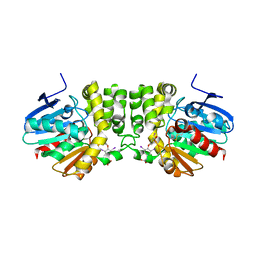 | |
3EN3
 
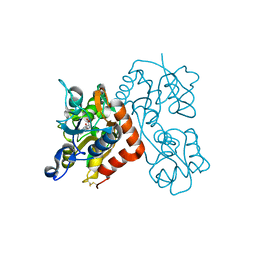 | | Crystal Structure of the GluR4 Ligand-Binding domain in complex with kainate | | Descriptor: | 3-(CARBOXYMETHYL)-4-ISOPROPENYLPROLINE, Glutamate receptor 4,Glutamate receptor | | Authors: | Gill, A, Madden, D.R. | | Deposit date: | 2008-09-25 | | Release date: | 2009-05-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Correlating AMPA receptor activation and cleft closure across subunits: crystal structures of the GluR4 ligand-binding domain in complex with full and partial agonists
Biochemistry, 47, 2008
|
|
3EPE
 
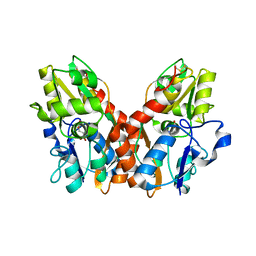 | |
8EE2
 
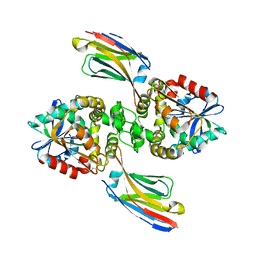 | |
8EVD
 
 | |
8E1C
 
 | |
8E1B
 
 | |
8E2N
 
 | |
1P0Z
 
 | | Sensor Kinase CitA binding domain | | Descriptor: | CITRATE ANION, MO(VI)(=O)(OH)2 CLUSTER, SODIUM ION, ... | | Authors: | Reinelt, S, Hofmann, E, Gerharz, T, Bott, M, Madden, D.R. | | Deposit date: | 2003-04-11 | | Release date: | 2003-08-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The structure of the periplasmic ligand-binding domain of the sensor kinase CitA reveals the first extracellular PAS domain.
J.Biol.Chem., 278, 2003
|
|
5HKB
 
 | |
5HKA
 
 | |
5HK9
 
 | |
5IC3
 
 | | CAL PDZ domain with peptide and inhibitor | | Descriptor: | 4-[methyl(nitroso)amino]benzene-1,2-diol, Golgi-associated PDZ and coiled-coil motif-containing protein, HPV18E6 Peptide | | Authors: | Zhao, Y, Madden, D.R. | | Deposit date: | 2016-02-22 | | Release date: | 2017-08-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | CAL PDZ domain with peptide and inhibitor
To Be Published
|
|
